Edge coloring with user data




str(ClusterList) List of 3 $ All : chr [1:1450] "89886" "29923" "100132891" "101410536" ... $ g1 : chr [1:858] "89886" "29923" "100132891" "101410536" ... $ g2: chr [1:592] "5325" "170691" "29953" "283392" ... CompareGO_BP=compareCluster(ClusterList, fun="enrichGO", pvalueCutoff=0.01, pAdjustMethod="BH", OrgDb=org.Hs.eg.db,ont="BP",readable=T) dotplot(CompareGO_BP, showCategory=10, title="GO - Biological Process")I ask for 10 categories, but ...
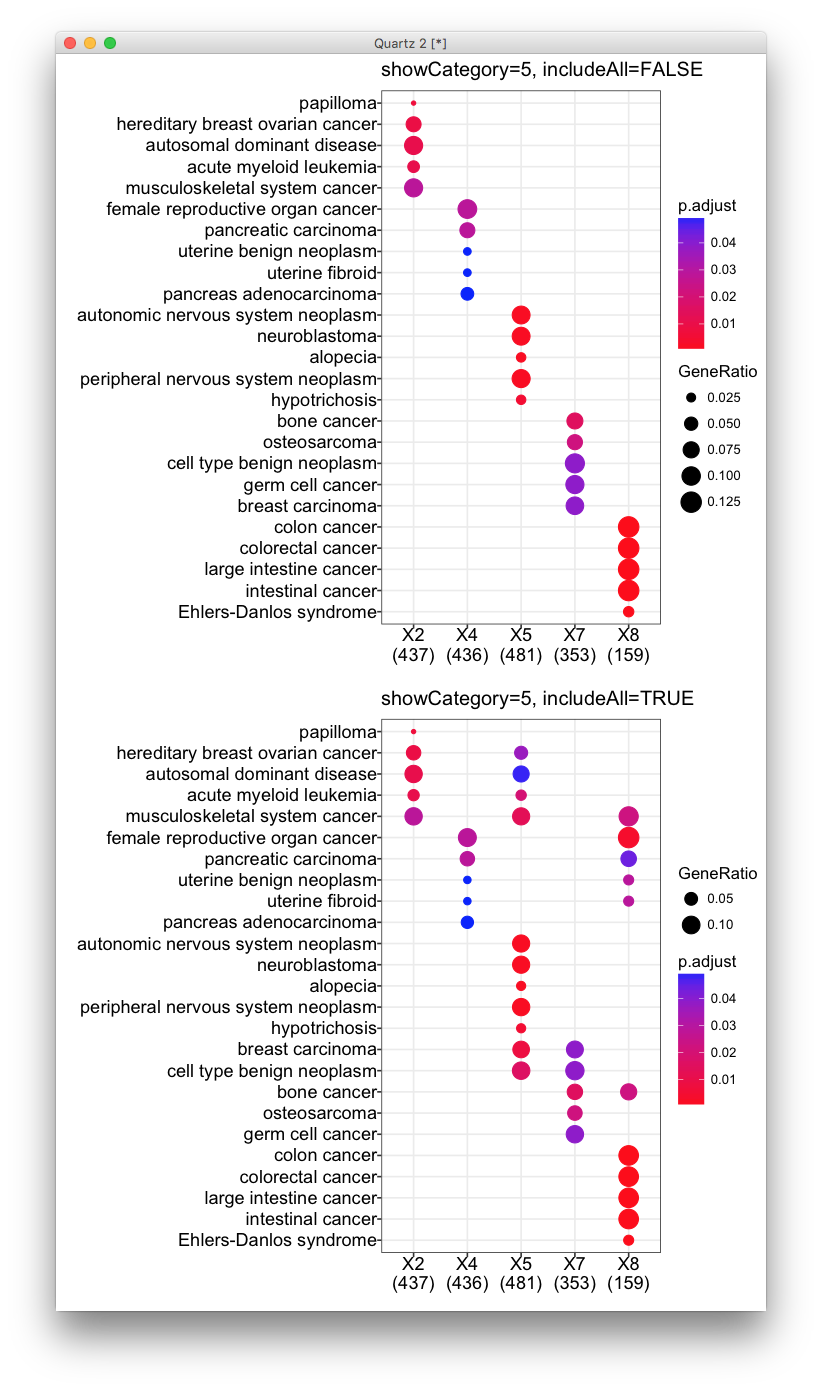
str(ClusterList) List of 3 $ All : chr [1:1450] "89886" "29923" "100132891" "101410536" ... $ g1 : chr [1:858] "89886" "29923" "100132891" "101410536" ... $ g2: chr [1:592] "5325" "170691" "29953" "283392" ... CompareGO_BP=compareCluster(ClusterList, fun="enrichGO", pvalueCutoff=0.01, pAdjustMethod="BH", OrgDb=org.Hs.eg.db,ont="BP",readable=T) dotplot(CompareGO_BP, showCategory=10, title="GO - Biological Process")I ask for 10 categories, but ...

CHANGES IN VERSION 1.9.8 ------------------------ o plotAvgProf/plotAvgProf2 order of panel by names of input tagMatrix List <2016-09-25, Sun> o test ENSEMBL ID using '^ENS' instead of '^ENSG' <2016-09-20, Tue> + https://github.com/GuangchuangYu/ChIPseeker/issues/41 CHANGES IN VERSION 1.9.7 ------------------------ o unit test <2016-08-16, Tue> CHANGES IN VERSION 1.9.6 ------------------------ o update vignette <2016-08-16, Tue> CHANGES IN VERSION 1.9.5 ------------------------ o when TxDb doesn't have gene_id information, converting gene ID (ensembl/entrez and symbol) will be omitted instead of throw error. <2016-08-02, Tue> + https://www.biostars.org/p/204142 o bug fixed if testing targetPeak is a list of GRanges objects in enrichPeakOverlap function <2016-07-20, Wed> + https://github.com/GuangchuangYu/ChIPseeker/issues/37 + https://github.com/GuangchuangYu/ChIPseeker/issues/36 o fixed typo in determine gene ID type <2016-06-21, Tue> + https://github.com/GuangchuangYu/ChIPseeker/issues/28#issuecomment-227212519 o move upsetplot generics to DOSE and import from DOSE to prevent function name conflict <2016-06-14, Tue> CHANGES IN VERSION 1.9.4 ------------------------ o bug fixed <2016-06-08, Wed> + https://github.com/GuangchuangYu/ChIPseeker/issues/17#issuecomment-224407402 + https://github.com/GuangchuangYu/ChIPseeker/pull/24/files CHANGES IN VERSION 1.9.3 ------------------------ o use byte compiler <2016-05-18, Wed> o https://github.com/Bioconductor-mirror/ChIPseeker/commit/f1ada57b9c66a1a44355bbbbdaf5b0a88e10cf7d CHANGES IN VERSION 1.9.2 ------------------------ o name tagMatrix in plotAvgProf automatically if missing <2016-05-12, Thu> o https://github.com/Bioconductor-mirror/ChIPseeker/commit/d5f16b2bc01725e30282c3acb33007ef521a514c CHANGES IN VERSION 1.9.1 ------------------------ o bug fixed in getNearestFeatureIndicesAndDistances <2016-05-11, Wed> + correct metadata in dummy NA featureclusterProfiler Thanks to the update of ... [Read more...]
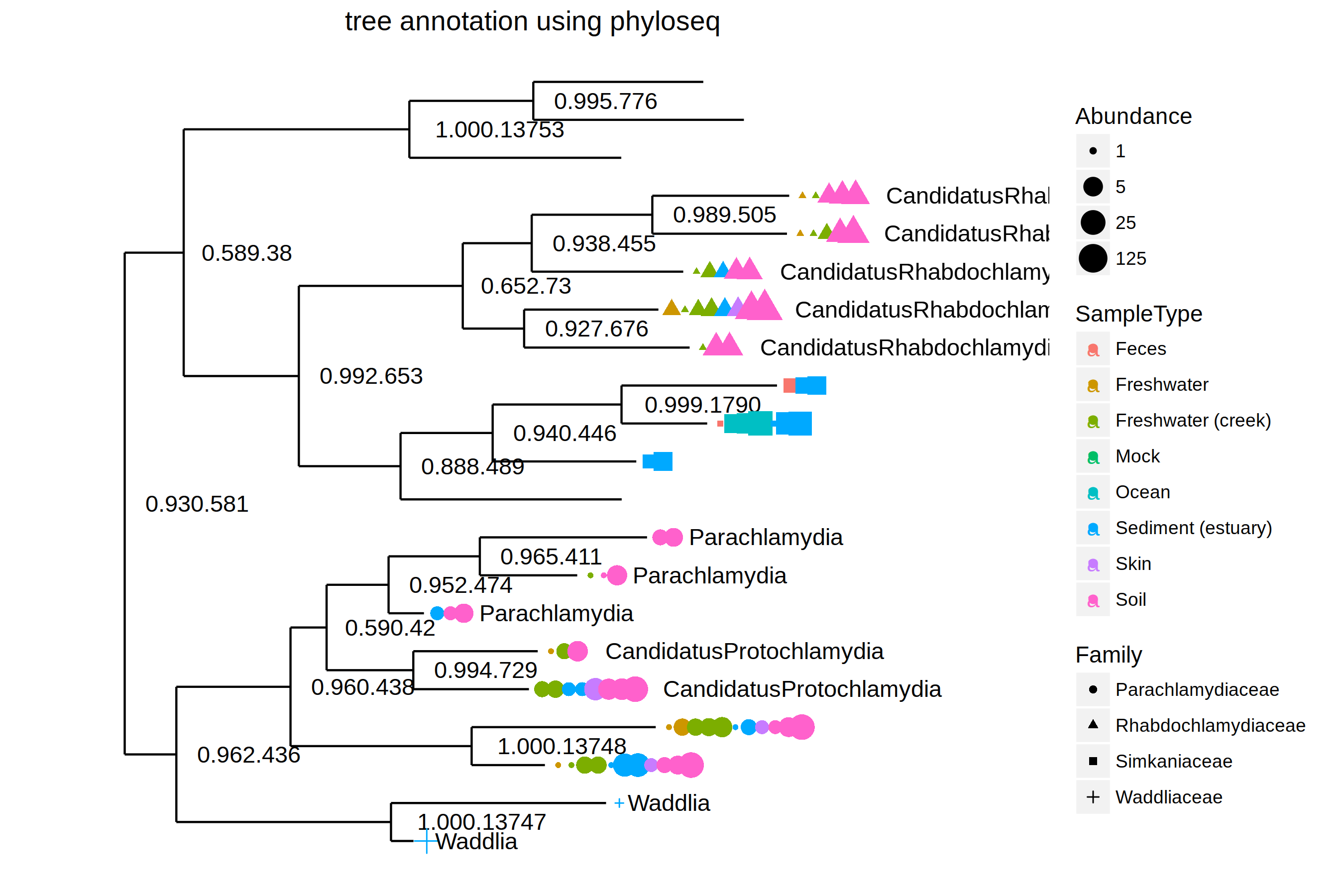
library(OutbreakTools)
data(FluH1N1pdm2009)
attach(FluH1N1pdm2009)
x <- new("obkData", individuals = individuals, dna = FluH1N1pdm2009$dna,
dna.individualID = samples$individualID, dna.date = samples$date,
trees = FluH1N1pdm2009$trees)
plotggphy(x, ladderize = TRUE, branch.unit = "year",
tip.color = "location", tip.size = 3, tip.alpha = 0.75)
As I mentioned in the post, ggtree for microbiome data, ggtree fits the R ecosystem ...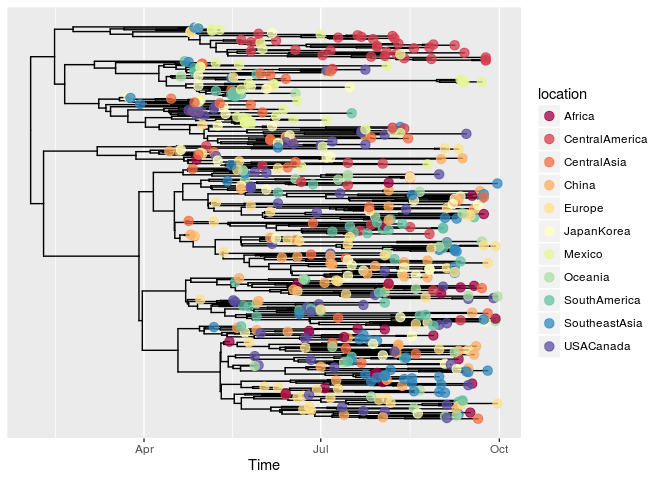


Copyright © 2025 | MH Corporate basic by MH Themes