Merging spatial buffers in R
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
I’m sure there’s a better way out there, but I struggled to find a way to dissolve polygons that touched/overlapped each other (the special case being buffers). For example, using the osmdata package, we can download the polygons representing hospital buildings in Bern, Switzerland.
library(osmdata) library(rgdal) ; library(maptools) ; library(rgeos) q0 <- opq(bbox = "Bern, Switzerland", timeout = 60) q1 <- add_osm_feature(q0, key = 'building', value = "hospital") x <- osmdata_sp(q1) library(leaflet) spChFIDs(x$osm_polygons) <- 1:nrow(x$osm_polygons@data) cent <- gCentroid(x$osm_polygons, byid = TRUE) leaflet(cent) %>% addTiles() %>% addCircles()
Here we plot the building centroids.
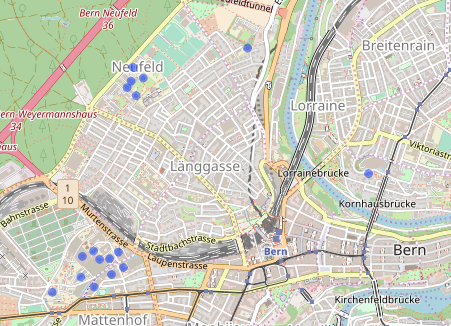
Each point represents a hospital building. We don’t particularly care about the buildings themselves though. We want to create hospitals. To do so, we try a 150m buffer around each centroid.
buff <- gBuffer(cent, byid = TRUE, width = 0.0015) leaflet(cent) %>% addTiles() %>% addPolygons(data = buff, col = "red") %>% addCircles()
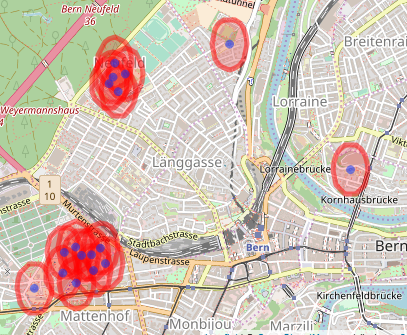
We then want to merge the buffers into, in this case, four groups. This is the point that doesn’t seem to be implemented anywhere that I could see (I also tried QGIS but that just created one big feature, rather than many small ones). My approach is to get the unique sets of intersections, add them as a variable to the buffer and unify the polygons.
buff <- SpatialPolygonsDataFrame(buff, data.frame(row.names = names(buff), n = 1:length(buff)))
gt <- gIntersects(buff, byid = TRUE, returnDense = FALSE)
ut <- unique(gt)
nth <- 1:length(ut)
buff$n <- 1:nrow(buff)
buff$nth <- NA
for(i in 1:length(ut)){
x <- ut[[i]]
buff$nth[x] <- i
}
buffdis <- gUnaryUnion(buff, buff$nth)
leaflet(cent) %>% addTiles() %>% addPolygons(data = buffdis, col = "red") %>% addCircles()
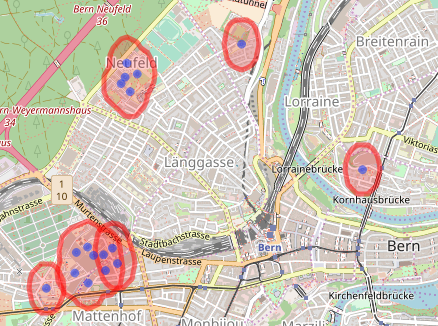
As you see, it almost worked. The lower left group is composed of three polygons. Doing the same process again clears it (only code shown). Large jobs might need more iterations (or larger buffers). The final job is to get the hospital centroids.
gt <- gIntersects(buffdis, byid = TRUE, returnDense = FALSE)
ut <- unique(gt)
nth <- 1:length(ut)
buffdis <- SpatialPolygonsDataFrame(buffdis, data.frame(row.names = names(buffdis), n = 1:length(buffdis)))
buffdis$nth <- NA
for(i in 1:length(ut)){
x <- ut[[i]]
buffdis$nth[x] <- i
}
buffdis <- gUnaryUnion(buffdis, buffdis$nth)
leaflet(cent) %>% addTiles() %>% addPolygons(data = buffdis, col = "red") %>% addCircles()
buffcent <- gCentroid(buffdis, byid = TRUE
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
