Practical Machine Learning with R and Python – Part 3
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
In this post ‘Practical Machine Learning with R and Python – Part 3’, I discuss ‘Feature Selection’ methods. This post is a continuation of my 2 earlier posts
- Practical Machine Learning with R and Python – Part 1
- Practical Machine Learning with R and Python – Part 2
While applying Machine Learning techniques, the data set will usually include a large number of predictors for a target variable. It is quite likely, that not all the predictors or feature variables will have an impact on the output. Hence it is becomes necessary to choose only those features which influence the output variable thus simplifying to a reduced feature set on which to train the ML model on. The techniques that are used are the following
- Best fit
- Forward fit
- Backward fit
- Ridge Regression or L2 regularization
- Lasso or L1 regularization
This post includes the equivalent ML code in R and Python.
All these methods remove those features which do not sufficiently influence the output. As in my previous 2 posts on “Practical Machine Learning with R and Python’, this post is largely based on the topics in the following 2 MOOC courses
1. Statistical Learning, Prof Trevor Hastie & Prof Robert Tibesherani, Online Stanford
2. Applied Machine Learning in Python Prof Kevyn-Collin Thomson, University Of Michigan, Coursera
You can download this R Markdown file and the associated data from Github – Machine Learning-RandPython-Part3.
1.1 Best Fit
For a dataset with features f1,f2,f3…fn, the ‘Best fit’ approach, chooses all possible combinations of features and creates separate ML models for each of the different combinations. The best fit algotithm then uses some filtering criteria based on Adj Rsquared, Cp, BIC or AIC to pick out the best model among all models.
Since the Best Fit approach searches the entire solution space it is computationally infeasible. The number of models that have to be searched increase exponentially as the number of predictors increase. For ‘p’ predictors a total of ML models have to be searched. This can be shown as follows
There are ways to choose single feature ML models among ‘n’ features,
ways to choose 2 feature models among ‘n’ models and so on, or
= Total number of models in Best Fit. Since from Binomial theorem we have
When x=1 in the equation (1) above, this becomes
Hence there are models to search amongst in Best Fit. For 10 features this is
or ~1000 models and for 40 features this becomes
which almost 1 trillion. Usually there are datasets with 1000 or maybe even 100000 features and Best fit becomes computationally infeasible.
Anyways I have included the Best Fit approach as I use the Boston crime datasets which is available both the MASS package in R and Sklearn in Python and it has 13 features. Even this small feature set takes a bit of time since the Best fit needs to search among ~ models
Initially I perform a simple Linear Regression Fit to estimate the features that are statistically insignificant. By looking at the p-values of the features it can be seen that ‘indus’ and ‘age’ features have high p-values and are not significant
1.1a Linear Regression – R code
source('RFunctions-1.R')
#Read the Boston crime data
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
# Rename the columns
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select specific columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
dim(df1)
## [1] 506 14
# Linear Regression fit
fit <- lm(cost~. ,data=df1)
summary(fit)
##
## Call:
## lm(formula = cost ~ ., data = df1)
##
## Residuals:
## Min 1Q Median 3Q Max
## -15.595 -2.730 -0.518 1.777 26.199
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3.646e+01 5.103e+00 7.144 3.28e-12 ***
## crimeRate -1.080e-01 3.286e-02 -3.287 0.001087 **
## zone 4.642e-02 1.373e-02 3.382 0.000778 ***
## indus 2.056e-02 6.150e-02 0.334 0.738288
## charles 2.687e+00 8.616e-01 3.118 0.001925 **
## nox -1.777e+01 3.820e+00 -4.651 4.25e-06 ***
## rooms 3.810e+00 4.179e-01 9.116 < 2e-16 ***
## age 6.922e-04 1.321e-02 0.052 0.958229
## distances -1.476e+00 1.995e-01 -7.398 6.01e-13 ***
## highways 3.060e-01 6.635e-02 4.613 5.07e-06 ***
## tax -1.233e-02 3.760e-03 -3.280 0.001112 **
## teacherRatio -9.527e-01 1.308e-01 -7.283 1.31e-12 ***
## color 9.312e-03 2.686e-03 3.467 0.000573 ***
## status -5.248e-01 5.072e-02 -10.347 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.745 on 492 degrees of freedom
## Multiple R-squared: 0.7406, Adjusted R-squared: 0.7338
## F-statistic: 108.1 on 13 and 492 DF, p-value: < 2.2e-16
Next we apply the different feature selection models to automatically remove features that are not significant below
1.1a Best Fit – R code
The Best Fit requires the ‘leaps’ R package
library(leaps)
source('RFunctions-1.R')
#Read the Boston crime data
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
# Rename the columns
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select specific columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Perform a best fit
bestFit=regsubsets(cost~.,df1,nvmax=13)
# Generate a summary of the fit
bfSummary=summary(bestFit)
# Plot the Residual Sum of Squares vs number of variables
plot(bfSummary$rss,xlab="Number of Variables",ylab="RSS",type="l",main="Best fit RSS vs No of features")
# Get the index of the minimum value
a=which.min(bfSummary$rss)
# Mark this in red
points(a,bfSummary$rss[a],col="red",cex=2,pch=20)
The plot below shows that the Best fit occurs with all 13 features included. Notice that there is no significant change in RSS from 11 features onward.

# Plot the CP statistic vs Number of variables plot(bfSummary$cp,xlab="Number of Variables",ylab="Cp",type='l',main="Best fit Cp vs No of features") # Find the lowest CP value b=which.min(bfSummary$cp) # Mark this in red points(b,bfSummary$cp[b],col="red",cex=2,pch=20)
Based on Cp metric the best fit occurs at 11 features as seen below. The values of the coefficients are also included below

# Display the set of features which provide the best fit coef(bestFit,b) ## (Intercept) crimeRate zone charles nox ## 36.341145004 -0.108413345 0.045844929 2.718716303 -17.376023429 ## rooms distances highways tax teacherRatio ## 3.801578840 -1.492711460 0.299608454 -0.011777973 -0.946524570 ## color status ## 0.009290845 -0.522553457 # Plot the BIC value plot(bfSummary$bic,xlab="Number of Variables",ylab="BIC",type='l',main="Best fit BIC vs No of Features") # Find and mark the min value c=which.min(bfSummary$bic) points(c,bfSummary$bic[c],col="red",cex=2,pch=20)

# R has some other good plots for best fit plot(bestFit,scale="r2",main="Rsquared vs No Features")
R has the following set of really nice visualizations. The plot below shows the Rsquared for a set of predictor variables. It can be seen when Rsquared starts at 0.74- indus, charles and age have not been included.

plot(bestFit,scale="Cp",main="Cp vs NoFeatures")
The Cp plot below for value shows indus, charles and age as not included in the Best fit

plot(bestFit,scale="bic",main="BIC vs Features")

1.1b Best fit (Exhaustive Search ) – Python code
The Python package for performing a Best Fit is the Exhaustive Feature Selector EFS.
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from mlxtend.feature_selection import ExhaustiveFeatureSelector as EFS
# Read the Boston crime data
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
# Set X and y
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
# Perform an Exhaustive Search. The EFS and SFS packages use 'neg_mean_squared_error'. The 'mean_squared_error' seems to have been deprecated. I think this is just the MSE with the a negative sign.
lr = LinearRegression()
efs1 = EFS(lr,
min_features=1,
max_features=13,
scoring='neg_mean_squared_error',
print_progress=True,
cv=5)
# Create a efs fit
efs1 = efs1.fit(X.as_matrix(), y.as_matrix())
print('Best negtive mean squared error: %.2f' % efs1.best_score_)
## Print the IDX of the best features
print('Best subset:', efs1.best_idx_)
Features: 8191/8191Best negtive mean squared error: -28.92
## ('Best subset:', (0, 1, 4, 6, 7, 8, 9, 10, 11, 12))
The indices for the best subset are shown above.
1.2 Forward fit
Forward fit is a greedy algorithm that tries to optimize the feature selected, by minimizing the selection criteria (adj Rsqaured, Cp, AIC or BIC) at every step. For a dataset with features f1,f2,f3…fn, the forward fit starts with the NULL set. It then pick the ML model with a single feature from n features which has the highest adj Rsquared, or minimum Cp, BIC or some such criteria. After picking the 1 feature from n which satisfies the criteria the most, the next feature from the remaining n-1 features is chosen. When the 2 feature model which satisfies the selection criteria the best is chosen, another feature from the remaining n-2 features are added and so on. The forward fit is a sub-optimal algorithm. There is no guarantee that the final list of features chosen will be the best among the lot. The computation required for this is of which is of the order of
. Though forward fit is a sub optimal solution it is far more computationally efficient than best fit
1.2a Forward fit – R code
Forward fit in R determines that 11 features are required for the best fit. The features are shown below
library(leaps)
# Read the data
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
# Rename the columns
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
#Split as training and test
train_idx <- trainTestSplit(df1,trainPercent=75,seed=5)
train <- df1[train_idx, ]
test <- df1[-train_idx, ]
# Find the best forward fit
fitFwd=regsubsets(cost~.,data=train,nvmax=13,method="forward")
# Compute the MSE
valErrors=rep(NA,13)
test.mat=model.matrix(cost~.,data=test)
for(i in 1:13){
coefi=coef(fitFwd,id=i)
pred=test.mat[,names(coefi)]%*%coefi
valErrors[i]=mean((test$cost-pred)^2)
}
# Plot the Residual Sum of Squares
plot(valErrors,xlab="Number of Variables",ylab="Validation Error",type="l",main="Forward fit RSS vs No of features")
# Gives the index of the minimum value
a<-which.min(valErrors)
print(a)
## [1] 11
# Highlight the smallest value
points(c,valErrors[a],col="blue",cex=2,pch=20)
Forward fit R selects 11 predictors as the best ML model to predict the ‘cost’ output variable. The values for these 11 predictors are included below

#Print the 11 ccoefficients coefi=coef(fitFwd,id=i) coefi ## (Intercept) crimeRate zone indus charles ## 2.397179e+01 -1.026463e-01 3.118923e-02 1.154235e-04 3.512922e+00 ## nox rooms age distances highways ## -1.511123e+01 4.945078e+00 -1.513220e-02 -1.307017e+00 2.712534e-01 ## tax teacherRatio color status ## -1.330709e-02 -8.182683e-01 1.143835e-02 -3.750928e-01
1.2b Forward fit with Cross Validation – R code
The Python package SFS includes N Fold Cross Validation errors for forward and backward fit so I decided to add this code to R. This is not available in the ‘leaps’ R package, however the implementation is quite simple. Another implementation is also available at Statistical Learning, Prof Trevor Hastie & Prof Robert Tibesherani, Online Stanford 2.
library(dplyr)
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
set.seed(6)
# Set max number of features
nvmax<-13
cvError <- NULL
# Loop through each features
for(i in 1:nvmax){
# Set no of folds
noFolds=5
# Create the rows which fall into different folds from 1..noFolds
folds = sample(1:noFolds, nrow(df1), replace=TRUE)
cv<-0
# Loop through the folds
for(j in 1:noFolds){
# The training is all rows for which the row is != j (k-1 folds -> training)
train <- df1[folds!=j,]
# The rows which have j as the index become the test set
test <- df1[folds==j,]
# Create a forward fitting model for this
fitFwd=regsubsets(cost~.,data=train,nvmax=13,method="forward")
# Select the number of features and get the feature coefficients
coefi=coef(fitFwd,id=i)
#Get the value of the test data
test.mat=model.matrix(cost~.,data=test)
# Multiply the tes data with teh fitted coefficients to get the predicted value
# pred = b0 + b1x1+b2x2... b13x13
pred=test.mat[,names(coefi)]%*%coefi
# Compute mean squared error
rss=mean((test$cost - pred)^2)
# Add all the Cross Validation errors
cv=cv+rss
}
# Compute the average of MSE for K folds for number of features 'i'
cvError[i]=cv/noFolds
}
a <- seq(1,13)
d <- as.data.frame(t(rbind(a,cvError)))
names(d) <- c("Features","CVError")
#Plot the CV Error vs No of Features
ggplot(d,aes(x=Features,y=CVError),color="blue") + geom_point() + geom_line(color="blue") +
xlab("No of features") + ylab("Cross Validation Error") +
ggtitle("Forward Selection - Cross Valdation Error vs No of Features")
Forward fit with 5 fold cross validation indicates that all 13 features are required

# This gives the index of the minimum value a=which.min(cvError) print(a) ## [1] 13 #Print the 13 coefficients of these features coefi=coef(fitFwd,id=a) coefi ## (Intercept) crimeRate zone indus charles ## 36.650645380 -0.107980979 0.056237669 0.027016678 4.270631466 ## nox rooms age distances highways ## -19.000715500 3.714720418 0.019952654 -1.472533973 0.326758004 ## tax teacherRatio color status ## -0.011380750 -0.972862622 0.009549938 -0.582159093
1.2c Forward fit – Python code
The Backward Fit in Python uses the Sequential feature selection (SFS) package (SFS)(https://rasbt.github.io/mlxtend/user_guide/feature_selection/SequentialFeatureSelector/)
Note: The Cross validation error for SFS in Sklearn is negative, possibly because it computes the ‘neg_mean_squared_error’. The earlier ‘mean_squared_error’ in the package seems to have been deprecated. I have taken the -ve of this neg_mean_squared_error. I think this would give mean_squared_error.
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.datasets import load_boston
from mlxtend.plotting import plot_sequential_feature_selection as plot_sfs
import matplotlib.pyplot as plt
from mlxtend.feature_selection import SequentialFeatureSelector as SFS
from sklearn.linear_model import LinearRegression
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
lr = LinearRegression()
# Create a forward fit model
sfs = SFS(lr,
k_features=(1,13),
forward=True, # Forward fit
floating=False,
scoring='neg_mean_squared_error',
cv=5)
# Fit this on the data
sfs = sfs.fit(X.as_matrix(), y.as_matrix())
# Get all the details of the forward fits
a=sfs.get_metric_dict()
n=[]
o=[]
# Compute the mean cross validation scores
for i in np.arange(1,13):
n.append(-np.mean(a[i]['cv_scores']))
m=np.arange(1,13)
# Get the index of the minimum CV score
# Plot the CV scores vs the number of features
fig1=plt.plot(m,n)
fig1=plt.title('Mean CV Scores vs No of features')
fig1.figure.savefig('fig1.png', bbox_inches='tight')
print(pd.DataFrame.from_dict(sfs.get_metric_dict(confidence_interval=0.90)).T)
idx = np.argmin(n)
print "No of features=",idx
#Get the features indices for the best forward fit and convert to list
b=list(a[idx]['feature_idx'])
print(b)
# Index the column names.
# Features from forward fit
print("Features selected in forward fit")
print(X.columns[b])
## avg_score ci_bound cv_scores \
## 1 -42.6185 19.0465 [-23.5582499971, -41.8215743748, -73.993608929...
## 2 -36.0651 16.3184 [-18.002498199, -40.1507894517, -56.5286659068...
## 3 -34.1001 20.87 [-9.43012884381, -25.9584955394, -36.184188174...
## 4 -33.7681 20.1638 [-8.86076528781, -28.650217633, -35.7246353855...
## 5 -33.6392 20.5271 [-8.90807628524, -28.0684679108, -35.827463022...
## 6 -33.6276 19.0859 [-9.549485942, -30.9724602876, -32.6689523347,...
## 7 -32.4082 19.1455 [-10.0177149635, -28.3780298492, -30.926917231...
## 8 -32.3697 18.533 [-11.1431684243, -27.5765510172, -31.168994094...
## 9 -32.4016 21.5561 [-10.8972555995, -25.739780653, -30.1837430353...
## 10 -32.8504 22.6508 [-12.3909282079, -22.1533250755, -33.385407342...
## 11 -34.1065 24.7019 [-12.6429253721, -22.1676650245, -33.956999528...
## 12 -35.5814 25.693 [-12.7303397453, -25.0145323483, -34.211898373...
## 13 -37.1318 23.2657 [-12.4603005692, -26.0486211062, -33.074137979...
##
## feature_idx std_dev std_err
## 1 (12,) 18.9042 9.45212
## 2 (10, 12) 16.1965 8.09826
## 3 (10, 12, 5) 20.7142 10.3571
## 4 (10, 3, 12, 5) 20.0132 10.0066
## 5 (0, 10, 3, 12, 5) 20.3738 10.1869
## 6 (0, 3, 5, 7, 10, 12) 18.9433 9.47167
## 7 (0, 2, 3, 5, 7, 10, 12) 19.0026 9.50128
## 8 (0, 1, 2, 3, 5, 7, 10, 12) 18.3946 9.19731
## 9 (0, 1, 2, 3, 5, 7, 10, 11, 12) 21.3952 10.6976
## 10 (0, 1, 2, 3, 4, 5, 7, 10, 11, 12) 22.4816 11.2408
## 11 (0, 1, 2, 3, 4, 5, 6, 7, 10, 11, 12) 24.5175 12.2587
## 12 (0, 1, 2, 3, 4, 5, 6, 7, 9, 10, 11, 12) 25.5012 12.7506
## 13 (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12) 23.0919 11.546
## No of features= 7
## [0, 2, 3, 5, 7, 10, 12]
## #################################################################################
## Features selected in forward fit
## Index([u'crimeRate', u'indus', u'chasRiver', u'rooms', u'distances',
## u'teacherRatio', u'status'],
## dtype='object')
The table above shows the average score, 10 fold CV errors, the features included at every step, std. deviation and std. error

The above plot indicates that 8 features provide the lowest Mean CV error
1.3 Backward Fit
Backward fit belongs to the class of greedy algorithms which tries to optimize the feature set, by dropping a feature at every stage which results in the worst performance for a given criteria of Adj RSquared, Cp, BIC or AIC. For a dataset with features f1,f2,f3…fn, the backward fit starts with the all the features f1,f2.. fn to begin with. It then pick the ML model with a n-1 features by dropping the feature,, for e.g., the inclusion of which results in the worst performance in adj Rsquared, or minimum Cp, BIC or some such criteria. At every step 1 feature is dopped. There is no guarantee that the final list of features chosen will be the best among the lot. The computation required for this is of
which is of the order of
. Though backward fit is a sub optimal solution it is far more computationally efficient than best fit
1.3a Backward fit – R code
library(dplyr)
# Read the data
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
# Rename the columns
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
set.seed(6)
# Set max number of features
nvmax<-13
cvError <- NULL
# Loop through each features
for(i in 1:nvmax){
# Set no of folds
noFolds=5
# Create the rows which fall into different folds from 1..noFolds
folds = sample(1:noFolds, nrow(df1), replace=TRUE)
cv<-0
for(j in 1:noFolds){
# The training is all rows for which the row is != j
train <- df1[folds!=j,]
# The rows which have j as the index become the test set
test <- df1[folds==j,]
# Create a backward fitting model for this
fitFwd=regsubsets(cost~.,data=train,nvmax=13,method="backward")
# Select the number of features and get the feature coefficients
coefi=coef(fitFwd,id=i)
#Get the value of the test data
test.mat=model.matrix(cost~.,data=test)
# Multiply the tes data with teh fitted coefficients to get the predicted value
# pred = b0 + b1x1+b2x2... b13x13
pred=test.mat[,names(coefi)]%*%coefi
# Compute mean squared error
rss=mean((test$cost - pred)^2)
# Add the Residual sum of square
cv=cv+rss
}
# Compute the average of MSE for K folds for number of features 'i'
cvError[i]=cv/noFolds
}
a <- seq(1,13)
d <- as.data.frame(t(rbind(a,cvError)))
names(d) <- c("Features","CVError")
# Plot the Cross Validation Error vs Number of features
ggplot(d,aes(x=Features,y=CVError),color="blue") + geom_point() + geom_line(color="blue") +
xlab("No of features") + ylab("Cross Validation Error") +
ggtitle("Backward Selection - Cross Valdation Error vs No of Features")

# This gives the index of the minimum value a=which.min(cvError) print(a) ## [1] 13 #Print the 13 coefficients of these features coefi=coef(fitFwd,id=a) coefi ## (Intercept) crimeRate zone indus charles ## 36.650645380 -0.107980979 0.056237669 0.027016678 4.270631466 ## nox rooms age distances highways ## -19.000715500 3.714720418 0.019952654 -1.472533973 0.326758004 ## tax teacherRatio color status ## -0.011380750 -0.972862622 0.009549938 -0.582159093
Backward selection in R also indicates the 13 features and the corresponding coefficients as providing the best fit
1.3b Backward fit – Python code
The Backward Fit in Python uses the Sequential feature selection (SFS) package (SFS)(https://rasbt.github.io/mlxtend/user_guide/feature_selection/SequentialFeatureSelector/)
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from mlxtend.plotting import plot_sequential_feature_selection as plot_sfs
import matplotlib.pyplot as plt
from mlxtend.feature_selection import SequentialFeatureSelector as SFS
from sklearn.linear_model import LinearRegression
# Read the data
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
lr = LinearRegression()
# Create the SFS model
sfs = SFS(lr,
k_features=(1,13),
forward=False, # Backward
floating=False,
scoring='neg_mean_squared_error',
cv=5)
# Fit the model
sfs = sfs.fit(X.as_matrix(), y.as_matrix())
a=sfs.get_metric_dict()
n=[]
o=[]
# Compute the mean of the validation scores
for i in np.arange(1,13):
n.append(-np.mean(a[i]['cv_scores']))
m=np.arange(1,13)
# Plot the Validation scores vs number of features
fig2=plt.plot(m,n)
fig2=plt.title('Mean CV Scores vs No of features')
fig2.figure.savefig('fig2.png', bbox_inches='tight')
print(pd.DataFrame.from_dict(sfs.get_metric_dict(confidence_interval=0.90)).T)
# Get the index of minimum cross validation error
idx = np.argmin(n)
print "No of features=",idx
#Get the features indices for the best forward fit and convert to list
b=list(a[idx]['feature_idx'])
# Index the column names.
# Features from backward fit
print("Features selected in bacward fit")
print(X.columns[b])
## avg_score ci_bound cv_scores \
## 1 -42.6185 19.0465 [-23.5582499971, -41.8215743748, -73.993608929...
## 2 -36.0651 16.3184 [-18.002498199, -40.1507894517, -56.5286659068...
## 3 -35.4992 13.9619 [-17.2329292677, -44.4178648308, -51.633177846...
## 4 -33.463 12.4081 [-20.6415333292, -37.3247852146, -47.479302977...
## 5 -33.1038 10.6156 [-20.2872309863, -34.6367078466, -45.931870352...
## 6 -32.0638 10.0933 [-19.4463829372, -33.460638577, -42.726257249,...
## 7 -30.7133 9.23881 [-19.4425181917, -31.1742902259, -40.531266671...
## 8 -29.7432 9.84468 [-19.445277268, -30.0641187173, -40.2561247122...
## 9 -29.0878 9.45027 [-19.3545569877, -30.094768669, -39.7506036377...
## 10 -28.9225 9.39697 [-18.562171585, -29.968504938, -39.9586835965,...
## 11 -29.4301 10.8831 [-18.3346152225, -30.3312847532, -45.065432793...
## 12 -30.4589 11.1486 [-18.493389527, -35.0290639374, -45.1558231765...
## 13 -37.1318 23.2657 [-12.4603005692, -26.0486211062, -33.074137979...
##
## feature_idx std_dev std_err
## 1 (12,) 18.9042 9.45212
## 2 (10, 12) 16.1965 8.09826
## 3 (10, 12, 7) 13.8576 6.92881
## 4 (12, 10, 4, 7) 12.3154 6.15772
## 5 (4, 7, 8, 10, 12) 10.5363 5.26816
## 6 (4, 7, 8, 9, 10, 12) 10.0179 5.00896
## 7 (1, 4, 7, 8, 9, 10, 12) 9.16981 4.58491
## 8 (1, 4, 7, 8, 9, 10, 11, 12) 9.77116 4.88558
## 9 (0, 1, 4, 7, 8, 9, 10, 11, 12) 9.37969 4.68985
## 10 (0, 1, 4, 6, 7, 8, 9, 10, 11, 12) 9.3268 4.6634
## 11 (0, 1, 3, 4, 6, 7, 8, 9, 10, 11, 12) 10.8018 5.40092
## 12 (0, 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12) 11.0653 5.53265
## 13 (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12) 23.0919 11.546
## No of features= 9
## Features selected in bacward fit
## Index([u'crimeRate', u'zone', u'NO2', u'distances', u'idxHighways', u'taxRate',
## u'teacherRatio', u'color', u'status'],
## dtype='object')
The table above shows the average score, 10 fold CV errors, the features included at every step, std. deviation and std. error

Backward fit in Python indicate that 10 features provide the best fit
1.3c Sequential Floating Forward Selection (SFFS) – Python code
The Sequential Feature search also includes ‘floating’ variants which include or exclude features conditionally, once they were excluded or included. The SFFS can conditionally include features which were excluded from the previous step, if it results in a better fit. This option will tend to a better solution, than plain simple SFS. These variants are included below
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.datasets import load_boston
from mlxtend.plotting import plot_sequential_feature_selection as plot_sfs
import matplotlib.pyplot as plt
from mlxtend.feature_selection import SequentialFeatureSelector as SFS
from sklearn.linear_model import LinearRegression
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
lr = LinearRegression()
# Create the floating forward search
sffs = SFS(lr,
k_features=(1,13),
forward=True, # Forward
floating=True, #Floating
scoring='neg_mean_squared_error',
cv=5)
# Fit a model
sffs = sffs.fit(X.as_matrix(), y.as_matrix())
a=sffs.get_metric_dict()
n=[]
o=[]
# Compute mean validation scores
for i in np.arange(1,13):
n.append(-np.mean(a[i]['cv_scores']))
m=np.arange(1,13)
# Plot the cross validation score vs number of features
fig3=plt.plot(m,n)
fig3=plt.title('SFFS:Mean CV Scores vs No of features')
fig3.figure.savefig('fig3.png', bbox_inches='tight')
print(pd.DataFrame.from_dict(sffs.get_metric_dict(confidence_interval=0.90)).T)
# Get the index of the minimum CV score
idx = np.argmin(n)
print "No of features=",idx
#Get the features indices for the best forward floating fit and convert to list
b=list(a[idx]['feature_idx'])
print(b)
print("#################################################################################")
# Index the column names.
# Features from forward fit
print("Features selected in forward fit")
print(X.columns[b])
## avg_score ci_bound cv_scores \
## 1 -42.6185 19.0465 [-23.5582499971, -41.8215743748, -73.993608929...
## 2 -36.0651 16.3184 [-18.002498199, -40.1507894517, -56.5286659068...
## 3 -34.1001 20.87 [-9.43012884381, -25.9584955394, -36.184188174...
## 4 -33.7681 20.1638 [-8.86076528781, -28.650217633, -35.7246353855...
## 5 -33.6392 20.5271 [-8.90807628524, -28.0684679108, -35.827463022...
## 6 -33.6276 19.0859 [-9.549485942, -30.9724602876, -32.6689523347,...
## 7 -32.1834 12.1001 [-17.9491036167, -39.6479234651, -45.470227740...
## 8 -32.0908 11.8179 [-17.4389015788, -41.2453629843, -44.247557798...
## 9 -31.0671 10.1581 [-17.2689542913, -37.4379370429, -41.366372300...
## 10 -28.9225 9.39697 [-18.562171585, -29.968504938, -39.9586835965,...
## 11 -29.4301 10.8831 [-18.3346152225, -30.3312847532, -45.065432793...
## 12 -30.4589 11.1486 [-18.493389527, -35.0290639374, -45.1558231765...
## 13 -37.1318 23.2657 [-12.4603005692, -26.0486211062, -33.074137979...
##
## feature_idx std_dev std_err
## 1 (12,) 18.9042 9.45212
## 2 (10, 12) 16.1965 8.09826
## 3 (10, 12, 5) 20.7142 10.3571
## 4 (10, 3, 12, 5) 20.0132 10.0066
## 5 (0, 10, 3, 12, 5) 20.3738 10.1869
## 6 (0, 3, 5, 7, 10, 12) 18.9433 9.47167
## 7 (0, 1, 2, 3, 7, 10, 12) 12.0097 6.00487
## 8 (0, 1, 2, 3, 7, 8, 10, 12) 11.7297 5.86484
## 9 (0, 1, 2, 3, 7, 8, 9, 10, 12) 10.0822 5.04111
## 10 (0, 1, 4, 6, 7, 8, 9, 10, 11, 12) 9.3268 4.6634
## 11 (0, 1, 3, 4, 6, 7, 8, 9, 10, 11, 12) 10.8018 5.40092
## 12 (0, 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12) 11.0653 5.53265
## 13 (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12) 23.0919 11.546
## No of features= 9
## [0, 1, 2, 3, 7, 8, 9, 10, 12]
## #################################################################################
## Features selected in forward fit
## Index([u'crimeRate', u'zone', u'indus', u'chasRiver', u'distances',
## u'idxHighways', u'taxRate', u'teacherRatio', u'status'],
## dtype='object')
The table above shows the average score, 10 fold CV errors, the features included at every step, std. deviation and std. error
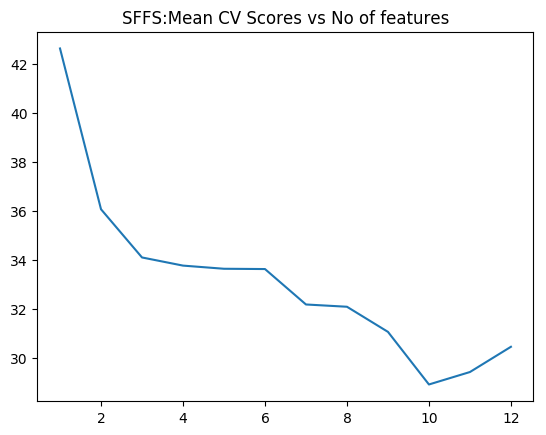
SFFS provides the best fit with 10 predictors
1.3d Sequential Floating Backward Selection (SFBS) – Python code
The SFBS is an extension of the SBS. Here features that are excluded at any stage can be conditionally included if the resulting feature set gives a better fit.
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.datasets import load_boston
from mlxtend.plotting import plot_sequential_feature_selection as plot_sfs
import matplotlib.pyplot as plt
from mlxtend.feature_selection import SequentialFeatureSelector as SFS
from sklearn.linear_model import LinearRegression
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
lr = LinearRegression()
sffs = SFS(lr,
k_features=(1,13),
forward=False, # Backward
floating=True, # Floating
scoring='neg_mean_squared_error',
cv=5)
sffs = sffs.fit(X.as_matrix(), y.as_matrix())
a=sffs.get_metric_dict()
n=[]
o=[]
# Compute the mean cross validation score
for i in np.arange(1,13):
n.append(-np.mean(a[i]['cv_scores']))
m=np.arange(1,13)
fig4=plt.plot(m,n)
fig4=plt.title('SFBS: Mean CV Scores vs No of features')
fig4.figure.savefig('fig4.png', bbox_inches='tight')
print(pd.DataFrame.from_dict(sffs.get_metric_dict(confidence_interval=0.90)).T)
# Get the index of the minimum CV score
idx = np.argmin(n)
print "No of features=",idx
#Get the features indices for the best backward floating fit and convert to list
b=list(a[idx]['feature_idx'])
print(b)
print("#################################################################################")
# Index the column names.
# Features from forward fit
print("Features selected in backward floating fit")
print(X.columns[b])
## avg_score ci_bound cv_scores \
## 1 -42.6185 19.0465 [-23.5582499971, -41.8215743748, -73.993608929...
## 2 -36.0651 16.3184 [-18.002498199, -40.1507894517, -56.5286659068...
## 3 -34.1001 20.87 [-9.43012884381, -25.9584955394, -36.184188174...
## 4 -33.463 12.4081 [-20.6415333292, -37.3247852146, -47.479302977...
## 5 -32.3699 11.2725 [-20.8771078371, -34.9825657934, -45.813447203...
## 6 -31.6742 11.2458 [-20.3082500364, -33.2288990522, -45.535507868...
## 7 -30.7133 9.23881 [-19.4425181917, -31.1742902259, -40.531266671...
## 8 -29.7432 9.84468 [-19.445277268, -30.0641187173, -40.2561247122...
## 9 -29.0878 9.45027 [-19.3545569877, -30.094768669, -39.7506036377...
## 10 -28.9225 9.39697 [-18.562171585, -29.968504938, -39.9586835965,...
## 11 -29.4301 10.8831 [-18.3346152225, -30.3312847532, -45.065432793...
## 12 -30.4589 11.1486 [-18.493389527, -35.0290639374, -45.1558231765...
## 13 -37.1318 23.2657 [-12.4603005692, -26.0486211062, -33.074137979...
##
## feature_idx std_dev std_err
## 1 (12,) 18.9042 9.45212
## 2 (10, 12) 16.1965 8.09826
## 3 (10, 12, 5) 20.7142 10.3571
## 4 (4, 10, 7, 12) 12.3154 6.15772
## 5 (12, 10, 4, 1, 7) 11.1883 5.59417
## 6 (4, 7, 8, 10, 11, 12) 11.1618 5.58088
## 7 (1, 4, 7, 8, 9, 10, 12) 9.16981 4.58491
## 8 (1, 4, 7, 8, 9, 10, 11, 12) 9.77116 4.88558
## 9 (0, 1, 4, 7, 8, 9, 10, 11, 12) 9.37969 4.68985
## 10 (0, 1, 4, 6, 7, 8, 9, 10, 11, 12) 9.3268 4.6634
## 11 (0, 1, 3, 4, 6, 7, 8, 9, 10, 11, 12) 10.8018 5.40092
## 12 (0, 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12) 11.0653 5.53265
## 13 (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12) 23.0919 11.546
## No of features= 9
## [0, 1, 4, 7, 8, 9, 10, 11, 12]
## #################################################################################
## Features selected in backward floating fit
## Index([u'crimeRate', u'zone', u'NO2', u'distances', u'idxHighways', u'taxRate',
## u'teacherRatio', u'color', u'status'],
## dtype='object')
The table above shows the average score, 10 fold CV errors, the features included at every step, std. deviation and std. error

SFBS indicates that 10 features are needed for the best fit
1.4 Ridge regression
In Linear Regression the Residual Sum of Squares (RSS) is given as
Ridge regularization =
where is the regularization or tuning parameter. Increasing increases the penalty on the coefficients thus shrinking them. However in Ridge Regression features that do not influence the target variable will shrink closer to zero but never become zero except for very large values of
Ridge regression in R requires the ‘glmnet’ package
1.4a Ridge Regression – R code
library(glmnet)
library(dplyr)
# Read the data
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
#Rename the columns
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Select specific columns
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Set X and y as matrices
X=as.matrix(df1[,1:13])
y=df1$cost
# Fit a Ridge model
fitRidge <-glmnet(X,y,alpha=0)
#Plot the model where the coefficient shrinkage is plotted vs log lambda
plot(fitRidge,xvar="lambda",label=TRUE,main= "Ridge regression coefficient shrikage vs log lambda")
The plot below shows how the 13 coefficients for the 13 predictors vary when lambda is increased. The x-axis includes log (lambda). We can see that increasing lambda from
to
significantly shrinks the coefficients. We can draw a vertical line from the x-axis and read the values of the 13 coefficients. Some of them will be close to zero

# Compute the cross validation error cvRidge=cv.glmnet(X,y,alpha=0) #Plot the cross validation error plot(cvRidge, main="Ridge regression Cross Validation Error (10 fold)")
This gives the 10 fold Cross Validation Error with respect to log (lambda) As lambda increase the MSE increases

1.4a Ridge Regression – Python code
The coefficient shrinkage for Python can be plotted like R using Least Angle Regression model a.k.a. LARS package. This is included below
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
from sklearn.linear_model import Ridge
X_train, X_test, y_train, y_test = train_test_split(X, y,
random_state = 0)
# Scale the X_train and X_test
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Fit a ridge regression with alpha=20
linridge = Ridge(alpha=20.0).fit(X_train_scaled, y_train)
# Print the training R squared
print('R-squared score (training): {:.3f}'
.format(linridge.score(X_train_scaled, y_train)))
# Print the test Rsquared
print('R-squared score (test): {:.3f}'
.format(linridge.score(X_test_scaled, y_test)))
print('Number of non-zero features: {}'
.format(np.sum(linridge.coef_ != 0)))
trainingRsquared=[]
testRsquared=[]
# Plot the effect of alpha on the test Rsquared
print('Ridge regression: effect of alpha regularization parameter\n')
# Choose a list of alpha values
for this_alpha in [0.001,.01,.1,0, 1, 10, 20, 50, 100, 1000]:
linridge = Ridge(alpha = this_alpha).fit(X_train_scaled, y_train)
# Compute training rsquared
r2_train = linridge.score(X_train_scaled, y_train)
# Compute test rsqaured
r2_test = linridge.score(X_test_scaled, y_test)
num_coeff_bigger = np.sum(abs(linridge.coef_) > 1.0)
trainingRsquared.append(r2_train)
testRsquared.append(r2_test)
# Create a dataframe
alpha=[0.001,.01,.1,0, 1, 10, 20, 50, 100, 1000]
trainingRsquared=pd.DataFrame(trainingRsquared,index=alpha)
testRsquared=pd.DataFrame(testRsquared,index=alpha)
# Plot training and test R squared as a function of alpha
df3=pd.concat([trainingRsquared,testRsquared],axis=1)
df3.columns=['trainingRsquared','testRsquared']
fig5=df3.plot()
fig5=plt.title('Ridge training and test squared error vs Alpha')
fig5.figure.savefig('fig5.png', bbox_inches='tight')
# Plot the coefficient shrinage using the LARS package
from sklearn import linear_model
# #############################################################################
# Compute paths
n_alphas = 200
alphas = np.logspace(0, 8, n_alphas)
coefs = []
for a in alphas:
ridge = linear_model.Ridge(alpha=a, fit_intercept=False)
ridge.fit(X_train_scaled, y_train)
coefs.append(ridge.coef_)
# #############################################################################
# Display results
ax = plt.gca()
fig6=ax.plot(alphas, coefs)
fig6=ax.set_xscale('log')
fig6=ax.set_xlim(ax.get_xlim()[::-1]) # reverse axis
fig6=plt.xlabel('alpha')
fig6=plt.ylabel('weights')
fig6=plt.title('Ridge coefficients as a function of the regularization')
fig6=plt.axis('tight')
plt.savefig('fig6.png', bbox_inches='tight')
## R-squared score (training): 0.620
## R-squared score (test): 0.438
## Number of non-zero features: 13
## Ridge regression: effect of alpha regularization parameter
The plot below shows the training and test error when increasing the tuning or regularization parameter ‘alpha’

For Python the coefficient shrinkage with LARS must be viewed from right to left, where you have increasing alpha. As alpha increases the coefficients shrink to 0.
1.5 Lasso regularization
The Lasso is another form of regularization, also known as L1 regularization. Unlike the Ridge Regression where the coefficients of features which do not influence the target tend to zero, in the lasso regualrization the coefficients become 0. The general form of Lasso is as follows
1.5a Lasso regularization – R code
library(glmnet)
library(dplyr)
df=read.csv("Boston.csv",stringsAsFactors = FALSE) # Data from MASS - SL
names(df) <-c("no","crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
df1 <- df %>% dplyr::select("crimeRate","zone","indus","charles","nox","rooms","age",
"distances","highways","tax","teacherRatio","color","status","cost")
# Set X and y as matrices
X=as.matrix(df1[,1:13])
y=df1$cost
# Fit the lasso model
fitLasso <- glmnet(X,y)
# Plot the coefficient shrinkage as a function of log(lambda)
plot(fitLasso,xvar="lambda",label=TRUE,main="Lasso regularization - Coefficient shrinkage vs log lambda")
The plot below shows that in L1 regularization the coefficients actually become zero with increasing lambda

# Compute the cross validation error (10 fold) cvLasso=cv.glmnet(X,y,alpha=0) # Plot the cross validation error plot(cvLasso)
This gives the MSE for the lasso model

1.5 b Lasso regularization – Python code
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso
from sklearn.preprocessing import MinMaxScaler
from sklearn import linear_model
scaler = MinMaxScaler()
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
X_train, X_test, y_train, y_test = train_test_split(X, y,
random_state = 0)
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
linlasso = Lasso(alpha=0.1, max_iter = 10).fit(X_train_scaled, y_train)
print('Non-zero features: {}'
.format(np.sum(linlasso.coef_ != 0)))
print('R-squared score (training): {:.3f}'
.format(linlasso.score(X_train_scaled, y_train)))
print('R-squared score (test): {:.3f}\n'
.format(linlasso.score(X_test_scaled, y_test)))
print('Features with non-zero weight (sorted by absolute magnitude):')
for e in sorted (list(zip(list(X), linlasso.coef_)),
key = lambda e: -abs(e[1])):
if e[1] != 0:
print('\t{}, {:.3f}'.format(e[0], e[1]))
print('Lasso regression: effect of alpha regularization\n\
parameter on number of features kept in final model\n')
trainingRsquared=[]
testRsquared=[]
#for alpha in [0.01,0.05,0.1, 1, 2, 3, 5, 10, 20, 50]:
for alpha in [0.01,0.07,0.05, 0.1, 1,2, 3, 5, 10]:
linlasso = Lasso(alpha, max_iter = 10000).fit(X_train_scaled, y_train)
r2_train = linlasso.score(X_train_scaled, y_train)
r2_test = linlasso.score(X_test_scaled, y_test)
trainingRsquared.append(r2_train)
testRsquared.append(r2_test)
alpha=[0.01,0.07,0.05, 0.1, 1,2, 3, 5, 10]
#alpha=[0.01,0.05,0.1, 1, 2, 3, 5, 10, 20, 50]
trainingRsquared=pd.DataFrame(trainingRsquared,index=alpha)
testRsquared=pd.DataFrame(testRsquared,index=alpha)
df3=pd.concat([trainingRsquared,testRsquared],axis=1)
df3.columns=['trainingRsquared','testRsquared']
fig7=df3.plot()
fig7=plt.title('LASSO training and test squared error vs Alpha')
fig7.figure.savefig('fig7.png', bbox_inches='tight')
## Non-zero features: 7
## R-squared score (training): 0.726
## R-squared score (test): 0.561
##
## Features with non-zero weight (sorted by absolute magnitude):
## status, -18.361
## rooms, 18.232
## teacherRatio, -8.628
## taxRate, -2.045
## color, 1.888
## chasRiver, 1.670
## distances, -0.529
## Lasso regression: effect of alpha regularization
## parameter on number of features kept in final model
##
## Computing regularization path using the LARS ...
## .C:\Users\Ganesh\ANACON~1\lib\site-packages\sklearn\linear_model\coordinate_descent.py:484: ConvergenceWarning: Objective did not converge. You might want to increase the number of iterations. Fitting data with very small alpha may cause precision problems.
## ConvergenceWarning)
The plot below gives the training and test R squared error

1.5c Lasso coefficient shrinkage – Python code
To plot the coefficient shrinkage for Lasso the Least Angle Regression model a.k.a. LARS package. This is shown below
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Lasso
from sklearn.preprocessing import MinMaxScaler
from sklearn import linear_model
scaler = MinMaxScaler()
df = pd.read_csv("Boston.csv",encoding = "ISO-8859-1")
#Rename the columns
df.columns=["no","crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status","cost"]
X=df[["crimeRate","zone","indus","chasRiver","NO2","rooms","age",
"distances","idxHighways","taxRate","teacherRatio","color","status"]]
y=df['cost']
X_train, X_test, y_train, y_test = train_test_split(X, y,
random_state = 0)
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
print("Computing regularization path using the LARS ...")
alphas, _, coefs = linear_model.lars_path(X_train_scaled, y_train, method='lasso', verbose=True)
xx = np.sum(np.abs(coefs.T), axis=1)
xx /= xx[-1]
fig8=plt.plot(xx, coefs.T)
ymin, ymax = plt.ylim()
fig8=plt.vlines(xx, ymin, ymax, linestyle='dashed')
fig8=plt.xlabel('|coef| / max|coef|')
fig8=plt.ylabel('Coefficients')
fig8=plt.title('LASSO Path - Coefficient Shrinkage vs L1')
fig8=plt.axis('tight')
plt.savefig('fig8.png', bbox_inches='tight')

1. Natural language processing: What would Shakespeare say?
2. Introducing QCSimulator: A 5-qubit quantum computing simulator in R
3. GooglyPlus: yorkr analyzes IPL players, teams, matches with plots and tables
4. My travels through the realms of Data Science, Machine Learning, Deep Learning and (AI)
5. Experiments with deblurring using OpenCV
6. R vs Python: Different similarities and similar differences
To see all posts see Index of posts
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
