UNHCR Refugee Data Visualized
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
I was working on to expand the disease transmission model from my first post but with the currents events, I felt compelled to work on something different. The new United States presidential administration formulated an executive order the end of last week, if issued, will suspend visas of internationals in the United States (either for school or work) from select countries, prevent the the entrance of Syrian refugees indefinitely and a “readjustment” of the the Refugee and Immigration Programs. Because of this disruptive change in immigration – especially for the most vulnerable populations that include refugees, asylum seekers and displaced people, all fleeing from conflict, instability or persecution – I wanted create a post on this topic.
Where’s the Data?
The data I’m using is taken from the United Nations High Commissioner for Refugees (UNHCR) website – the UN Refugee Agency. You can read more on what they do and why the exist in the link above. Currently you can only download the mid-year statistics for 2015. You get a statement saying: “Error: Statistics filters are not available at this time. Please try again later.” I just checked again and the data is accessible again!
In this post I am not going to share my views on the current political landscape in the United States (that can be a complete separate blog on its own). Similar to my previous post, I’m going to do a walkthrough with data cleaning, ask a couple questions and visualize the data in some meaningful way. If you want to skip the details on the code, you can just scroll down to figures.
Working in R
As usual, all the code is in my GitHub repository for whoever wants to download and use it for themselves. For this post, I’m going to use the following packages ggplot, grid, scales, reshape2, scales and worldcloud. I was reading some R manuals and I discovered a “new” way to access your working directory environment. It involves the use of the list.files() function, which lists the files or folders in your current working directory. You can also further specify the path name (which saves the time of constantly changing the pathname in the setwd() function (which I have been foolishly doing for a while).
setwd("~/Documents/UNHCR Data/") # ~ acts as base for home directory
list.files(full.names=TRUE) # gives full path for files or folders
files.all <- list.files(path="All_Data/") #assigns name to file names in the All_Data folder
length(files.all) #checks how many objects there are in the /All_Data folder
files.all
Since the file is a comm delimited file (.csv) we use the read.csv() function to read it in and assign it the name “ref.d”. I used what I know with the paste0() function We set the skip argument equal to 2 because by visual inspection of the file, the first two rows are committed to the file title (we don’t want to read that into R). I also used the na.string argument to specify that any blanks (“”), dashes (“-“) and asterisks (“*”) would be considered missing data, NA. The asterisks are specified to be redacted information, based on the UNHCR website.
ref.d <- read.csv(paste0("All_Data/",files.all), #insert filepath and name into 1st argument
header=T, #select the headers in 3rd row
skip=2, #skips the first rows (metadata in .csv file)
na.string=c("","-","*"), #convert all blanks, "i","*" cells into missing type NA
col.names=new.names #since we already made new names
)
First thing I see, the names for the columns names are long and they would be annoying to reproduce. So first we’ll change these using the names() function.
new.names <- c("Year", "Country", "Country_Origin", "Refugees", "Asylum_Seekers", "Returned_Refugees", "IDPs", "Returned_IDPs", "Stateless_People", "Others_of_Concern","Total")
names(ref.d) <- new.names
By using summary() and str() on the dataset we can see that the range of the data spans from 1951 to 2014, it contains information on the country where refugees are situated, their country of origin, counts for each population of concern and total counts.
> summary(ref.d)
Year Country Country_Origin Refugees
Min. :1951 Length:103746 Length:103746 Min. : 1
1st Qu.:2000 Class :character Class :character 1st Qu.: 3
Median :2006 Mode :character Mode :character Median : 16
Mean :2004 Mean : 5992
3rd Qu.:2010 3rd Qu.: 167
Max. :2014 Max. :3272290
NA's :19353
Asylum_Seekers Returned_Refugees IDPs Returned_IDPs
Min. : 0.0 Min. : 1 Min. : 470 Min. : 23
1st Qu.: 1.0 1st Qu.: 2 1st Qu.: 90746 1st Qu.: 5000
Median : 5.0 Median : 16 Median : 261704 Median : 27284
Mean : 255.9 Mean : 6737 Mean : 540579 Mean : 111224
3rd Qu.: 34.0 3rd Qu.: 247 3rd Qu.: 594443 3rd Qu.: 104230
Max. :358056.0 Max. :9799410 Max. :7632500 Max. :1186889
NA's :46690 NA's :97327 NA's :103330 NA's :103544
Stateless_People Others_of_Concern Total
Min. : 1 Min. : 1.0 Min. : 1
1st Qu.: 205 1st Qu.: 14.8 1st Qu.: 3
Median : 1720 Median : 444.5 Median : 16
Mean : 66803 Mean : 24672.5 Mean : 8605
3rd Qu.: 11462 3rd Qu.: 6000.0 3rd Qu.: 166
Max. :3500000 Max. :957000.0 Max. :9799410
NA's :103103 NA's :103018 NA's :2433
It’s always good practice to identify missing data as well (especially when we set the condition of the read.csv argument above). For non-numeric variables you can use a simple function using apply() and is.na() to identify missing values (NA’s) in your data.
apply(ref.d,2, function(x) sum(is.na(x)))
When I used str() on the data, I saw that the country names were as factors and the populations of concern categories were integers. I made a short for loop to change these to a character and numeric type respectively.
for(i in 2:length(names(ref.d))){ # "2"-ignores the first column (we want to keep Year as an integer)
if (class(ref.d[,i])=="factor"){
ref.d[,i] <- as.character(ref.d[,i])}
if (class(ref.d[,i])=="integer"){
ref.d[,i] <- as.numeric(ref.d[,i])}
}
Also another nuance, I wanted to change some of the names of the countries (they were either very long or had extra information). I first identified the names I wanted to change and then replace them a new set of names. I did this using a for loop as well.
old.countries <- c("Bolivia (Plurinational State of)",
"China, Hong Kong SAR",
"China, Macao SAR",
"Iran (Islamic Rep. of)",
"Micronesia (Federated States of)",
"Serbia and Kosovo (S/RES/1244 (1999))",
"Venezuela (Bolivarian Republic of)",
"Various/Unknown")
# replacement names
new.countries <- c("Bolivia","Hong Kong","Macao","Iran","Micronesia","Serbia & Kosovo","Venezuela","Unknown")
for (k in 1:length(old.countries)){
ref.d$Country_Origin[ref.d$Country_Origin==old.countries[k]]<-new.countries[k]
ref.d$Country[ref.d$Country==old.countries[k]]<-new.countries[k]
}
If any has alternative ways to achieve the above (ie. using the apply family), comment below! Just a short disclaimer on for loops in R. There has been a lot of argument on the effectiveness of for loops in R compared to the apply function family. A quick Google Search shows many opinions on this issue, based on computing speed/power, simplicity, elegance, etc. Advanced R by Hadley Wickham talks about this and I’ve generally used this as a guideline on whether to use a for loop or an apply function.
Some Descriptives and North Korea
Just to get an idea of the data, we can create a list of the countries and countries of origin by using the code similar to identifying countries with certain MRSA strains in my previous post.
clist<-sort(unique(ref.d$Country)) #alphabetical clist or.clist<-sort(unique(ref.d$Country_Origin)) #alphabetical or.clist
We can then compare them for any differences either using matching operators or the setdiff() function. First we’ll do this…
clist[!clist %in% or.clist] # or setdiff(clist,or.clist) [1] "Bonaire" "Montserrat" [3] "Sint Maarten (Dutch part)" "State of Palestine"
… we can infer that these countries haven’t produced refugees or there is no data on these countries in the UNHCR database. If we reverse the comparison…
or.clist[!or.clist %in% clist] # or setdiff(or.clist,clist) [1] "Andorra" "Anguilla" [3] "Bermuda" "Cook Islands" [5] "Dem. People's Rep. of Korea" "Dominica" [7] "French Polynesia" "Gibraltar" [9] "Guadeloupe" "Holy See (the)" [11] "Kiribati" "Maldives" [13] "Marshall Islands" "Martinique" [15] "New Caledonia" "Niue" [17] "Norfolk Island" "Palestinian" [19] "Puerto Rico" "Samoa" [21] "San Marino" "Sao Tome and Principe" [23] "Seychelles" "Stateless" [25] "Tibetan" "Tuvalu" [27] "Wallis and Futuna Islands " "Western Sahara"
… we get a list of countries that have only produced refugees (not taken any refugees in) or there is missing data in the UNHCR. I myself being Korean-Canadian I noticed North Korea (the Democratic People’s Republic of Korea) on this list. I wanted to ask the question, which countries have the largest number of North Korean refugees based on the UNHCR Data? What are the top 10?
NK.tot<- aggregate(cbind(Total)~Country,data=NK,FUN=sum)
NK.tot[order(-NK.tot[,2]),][1:10,]
Country Total
31 United Kingdom 4808
5 Canada 2954
10 Germany 2845
18 Netherlands 487
3 Belgium 435
23 Russian Federation 357
32 United States of America 346
1 Australia 318
20 Norway 262
9 France 228
We find that the UK has the highest number of North Koreans refugees, followed by Canada and Germany with similar counts. Learned something new today.
Word Clouds in R
If you haven’t guessed based on the packages I loaded in the beginning, a word cloud was inevitable. Making word clouds in R is pretty simple thanks to the wordcloud package. Remember that word clouds are an esthetically decent way to qualitatively see your data. There are bunch of pros and cons on using word clouds. Generally this is appropriate if you just want see the general relative frequency of words in your data. Making any quantitative conclusions would be erroneous.
First we’ll aggregate the counts for the country of origin.
or.count.tot <- aggregate(cbind(Total)~Country_Origin,data=ref.d,FUN=sum) or.count.tot
Then set color palette using HEX codes (much thanks to I Want Hue).
pal3 <- c("#274c56",
"#664c47",
"#4e5c48",
"#595668",
"#395e68",
"#516964",
"#6b6454",
"#58737f",
"#846b6b",
"#807288",
"#758997",
"#7e9283",
"#a79486",
"#aa95a2",
"#8ba7b4")
Then let’s make a word cloud based on the countries of origin for the populations of concern in the data and save it as a .png file. I’ve commented the code for each line We can see that Afghanistan seems to have the highest relative number and those with unknown countries of origin is second. Other high conflict countries are visible too like DRC, Iraq, Sudan, Syria, Somalia, etc.
# COUNTRY OF ORIGIN
png("wordcloud_count_or.png",width=600,height=850,res=200)
wordcloud(or.count.tot[,1], #list of words
or.count.tot[,2], #frequencies for words
scale=c(3,.5), #scale of size range
min.freq=100, #minimum frequency
max.words=100, #maximum number of words show (others dropped)
family="Garamond", font=2, #text edit (The font "face" (1=plain, 2=bold, 3=italic, 4=bold-italic))
random.order=F, #F-plotted in decreasing frequency
colors=rev(pal3)) #colours from least to most frequent (reverse order)
dev.off()
Historical Data Visualized
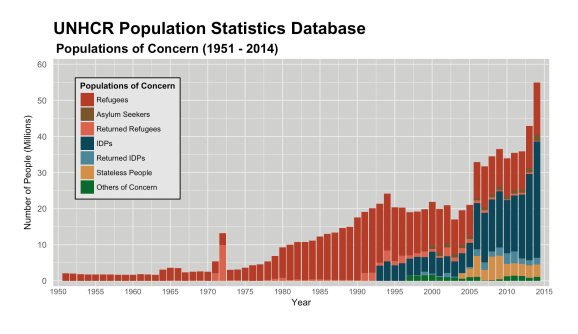
So in the UNHCR data, it contains information not only on countries but based on the year from 1951 to 2014. I wanted to see the trends over time between each of the populations of concern. There are seven types of populations in the dataset. Their definitions can be found here on the UNHCR website. To visualize the data using ggplot2, it’s a good practice to convert the data from wide to long form. The benefits on using long form can be read here and here. The accomplish this, we’re going to use the reshape2 package and the melt() function. This tutorial is helpful in understanding what’s going on.
I first subset to remove the country, country of origin and total columns.
no.count<-(ref.d[c(-2,-3,-11)]) #removes Country, Country of Origin and Total
lno.count<-melt(no.count,id=c("Year"))
head(lno.count)
Year variable value
1 1951 Refugees 180000
2 1951 Refugees 282000
3 1951 Refugees 55000
4 1951 Refugees 168511
5 1951 Refugees 10000
6 1951 Refugees 265000
Then I set the ggplot2 theme and the color palette I want to use.
blank_t <- theme_minimal()+
theme(
panel.border = element_blank(), #removes border
panel.background = element_rect(fill = "#d0d1cf",colour=NA),
panel.grid = element_line(colour = "#ffffff"),
plot.title=element_text(size=20, face="bold",hjust=0,vjust=2), #set the plot title
plot.subtitle=element_text(size=15,face=c("bold","italic"),hjust=0.01), #set subtitle
legend.title=element_text(size=10,face="bold",vjust=0.5), #specify legend title
axis.text.x = element_text(size=9,angle = 0, hjust = 0.5,vjust=1,margin=margin(r=10)),
axis.text.y = element_text(size=10,margin = margin(r=5)),
legend.background = element_rect(fill="gray90", size=.5),
legend.position=c(0.15,0.65),
plot.margin=unit(c(t=1,r=1.2,b=1.2,l=1),"cm")
)
pal5 <- c("#B53C26", #Refugees 7 TOTAL
"#79542D", #Asylum Seekers
"#DA634D", #Returned Refugees
"#0B4659", #IDPs
"#4B8699", #Returned IDPs
"#D38F47", #Stateless People
"#09692A") #Others of Concern
Then use geom_bar and the fill argument to make the stacked bar graph over time by specifying what titles and axis labels. In the scale_fill_manual() I used the gsub() function remove the underscores from the names (I was lazy to rewrite the names out in a vector and assign it to the labels).
gg1 <-ggplot(lno.count,aes(Year,value)) +
geom_bar(aes(fill=variable),stat="identity") +
labs(title="UNHCR Population Statistics Database",
subtitle="(1951 - 2014)",
x="Year",y="Number of People (Millions)") + blank_t +
scale_fill_manual(guide_legend(title="Populations of Concern"),labels=gsub("*_"," ",names(ref.d)[c(-1,-2,-3,-11)]),values=pal5)
Next while modifying the axes, I wanted to change the scale of the y-axis to shows it as millions versus 6 zeros. I set the labels argument in scale_y_continuous() to a short function that converts it by a factor of 10e-6. The plot is below and you can click on it to see the full-sized version.
mil.func gg2<-gg1+ scale_y_continuous(limits=c(0,6e7), breaks=pretty(0:6e7,n=5), labels=mil.func, expand=c(0.025,0)) + scale_x_continuous(limits=c(1950,2015), breaks=seq(1950,2015,by=5), expand=c(0.01,0)) ggsave(plot=gg2,filename="UNHCR_Totals_Yr.png", width=9.5, height=5.5, dpi=200)
So based on this, we can see that there has been a large increase of the total populations of concern the past 30 years. Internally Displaced People (IDPs) have also increased dramatically in the past 20 years. Based on the most recent data, the totals are reaching 65.3 million! (that’s above the y-limit in the bar graph)
Let’s Be Real
(Not R related) Based on doing this, I learned that the UK hosts the highest number of North Korean refugees in the world, that Afghanistan has historically produced the largest number of refugees and there is a upward trend (qualitative conclusion) with the total number of displaced people in recent history. Remembering that these vulnerable populations are mostly undocumented so the numbers in this dataset are likely underestimates to the actual numbers of people who fall under the population of concern definition. In the midst of the news and social media firestorm on all the unfortunate events happening in the world, I think it’s easy to dissociate and just live our own lives. I believe that those with access to resources should be generous with how we use them. A simple act of kindness like a Samaritan can go a long way. Some simple steps that I’ve taken were to educate myself on these issues, advocate where I am able, donate financially and engage where I can. There are a plethora of organizations committed to relieving the burden of these populations – one of them I support is the International Rescue Committee.
So I hope this post was informative and demonstrates how one can pull some data off the internet, tinker around with R and discover some news things about the world. Any feedback on the code, alternative ways on what I did are always welcome!
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.