Size of XDF files using RevoScaleR package
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
It came to my attention that size of XDF (external data frame) file can change drastically based on the compute context and environment. When testing the output of a dataset I was working on in SQL Server Management Studio I was simultaneously testing R code in RTVS or R Studio and I have noticed a file growth.
Following stored procedure will do a simple test:
CREATE PROCEDURE rxImport_Test ( @rowsPerRead INT )
AS
BEGIN
DECLARE @RStat NVARCHAR(4000)
SET @RStat = 'library(RevoScaleR)
#rxSetComputeContext("RxLocalSeq")
ptm <- proc.time()
inFile <- file.path(rxGetOption("sampleDataDir"), "AirlineDemoSmall.csv")
filename <- "AirlineDemoSmall_'+CAST(@rowsPerRead AS VARCHAR(100))+'_TSQL_NC.xdf"
rxTextToXdf(inFile = inFile, outFile = filename, stringsAsFactors = T, rowsPerRead = '+CAST(@rowsPerRead AS VARCHAR(100))+', overwrite=TRUE)
outFile <- file.path(rxGetOption("sampleDataDir"), filename)
rxImport(inData = inFile, outFile = outFile, overwrite=TRUE)
d <- proc.time() - ptm
filesize <- data.frame(file.size(filename))
time <- data.frame(d[3])
RowsPerRead <- data.frame('+CAST(@rowsPerRead AS VARCHAR(100))+')
filename_xdf <- data.frame(filename)
ran <- data.frame(Sys.time())
OutputDataSet <- cbind(as.character(filesize), time, RowsPerRead, filename_xdf, ran)';
EXECUTE sp_execute_external_script
@language = N'R'
,@script = @RStat
WITH RESULT SETS ((
Filesize NVARCHAR(100)
,Time_df NVARCHAR(100)
,RowsPerRead NVARCHAR(100)
,filename_xdf NVARCHAR(100)
,DateExecute NVARCHAR(100)
))
END
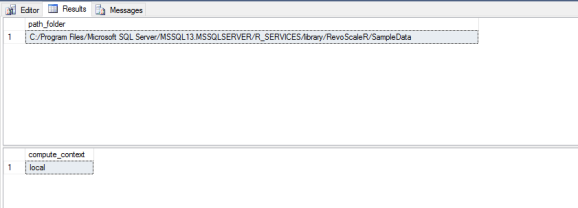
But let’s first understand and test the Computation context and path to the data.
-- Make sure your path location is pointing to RevoscaleR library folder!
EXECUTE sp_execute_external_script
@language = N'R'
,@script = N'library(RevoScaleR)
OutputDataSet <- data.frame(rxGetOption("sampleDataDir"))'
WITH RESULT SETS ((
path_folder NVARCHAR(1000)
))
-- check for ComputeContext
DECLARE @RStat NVARCHAR(4000)
SET @RStat = 'library(RevoScaleR)
cc <- rxGetOption("computeContext")
OutputDataSet <- data.frame(cc@description)';
EXECUTE sp_execute_external_script
@language = N'R'
,@script = @RStat
WITH RESULT SETS ((compute_context NVARCHAR(100)))
At my computer, this looks like this:
No we will run procedure
rxImport_Test
with different chunk sizes (this is what I will test) and observe execution times.
INSERT INTO rxImport_results EXEC rxImport_Test @rowsPerRead = 2; GO INSERT INTO rxImport_results EXEC rxImport_Test @rowsPerRead = 20; GO INSERT INTO rxImport_results EXEC rxImport_Test @rowsPerRead = 200; GO INSERT INTO rxImport_results EXEC rxImport_Test @rowsPerRead = 2000; GO
Running with different chunk size the procedure, it yields interesting results:

Now, let’s see the summary information on this file / dataset.
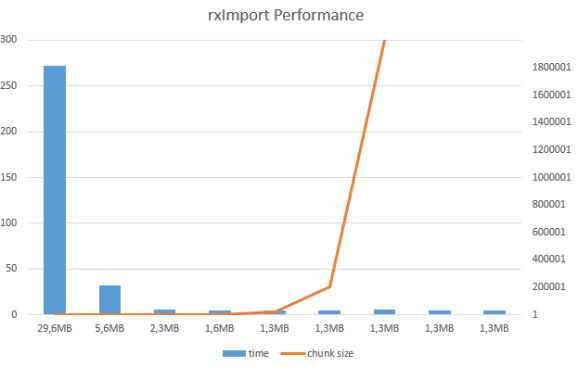
Considering that original file holds 600.000 rows, number of blocks for each of the files is also changing and therefore the size of the files is growing.
Retrieving information on block size
DECLARE @RStat NVARCHAR(4000) SET @RStat = 'library(RevoScaleR) info <- rxGetInfoXdf(data="AirlineDemoSmall_20000000_TSQL_NC.xdf", getVarInfo = TRUE) OutputDataSet <- data.frame(info$numBlocks)'; EXECUTE sp_execute_external_script @language = N'R' ,@script = @RStat WITH RESULT SETS (( nof_blocks NVARCHAR(100)))
one can see the change between the files and where is the optimal block size. In my test, number of blocks would be 3 to 30 max to receive maximum performance from creating XDF file. This means from 2000 up to 200.000 rows per block would yield best performance results. Otherwise I haven’t found the the golden rule of the block size, but take caution, especially when dealing with larger files.
I ran test couple of times in order to check the consistency of the results, and they hold water. As for the file size; this is the presentation of internal file, as of *.xdf file (as external structure) size should not differ as the block size changes, but perfomance does!
Code is available at Github.
Happy R-sqling!
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
