Calculates population growth rate λ along element changes
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
The previous article introduced the sensitivity and elasticity to seasonal matrix model of imaginary annual plant. Both sensitivity and elasticity are partial derivatives. This means the values can only predict a change of λ with respect to a small change of a element.
To know how λ will affected by a large shift or changes of multiple elements, the simplest way is to calculate each λ for each case.
R can easily do this.
The λ can also be solved analytically, because this example is very simple. Let’s check whether both results match.
We have four elements:
seed <- 0.9^4 # Seed surviving rate; annual germ <- 0.3 # Germination rate; spring plant <- 0.05 # Plant surviving rate; from germination to mature yield <- 100 # Seed production number; per matured plant
The function lambda and A.spring were defined in the previous article:
lambda <- function(A) eigen(A)$values[1] # and so on.
Let’s change one of them; the seed:
n <- 100
lambdas <- numeric(n)
seeds <- seq(from=0, to=1, length.out=n)^2
for(i in 1:n) {
seed <- seeds[i]
lambdas[i] <- lambda(A.spring())
}
seed <- 0.9^4 # restore the initial value
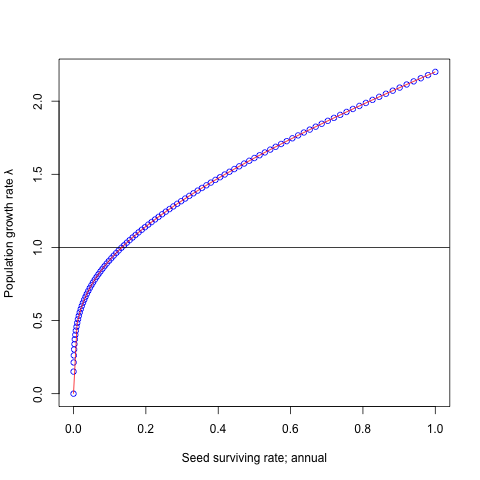
plot(seeds, lambdas, ylab='Population growth rate λ',
xlab='Seed surviving rate; annual', col='blue')
abline(a=1, b=0)
Blue plots indicate the result of simulation.
From analytic solution:
# λ = 0.7 * seed + 1.5 * seed^(1/4)
Drawing this curve with red line on the blue plots.
curve(0.7 * x + 1.5 * x^(1/4), add=T,
from=min(seeds), to=max(seeds), col='red')
Both results met very well.
The germ:
germs <- seq(from=0, to=1, length.out=n)
for(i in 1:n) {
germ <- germs[i]
lambdas[i] <- lambda(A.spring())
}
germ <- 0.3 # restore the initial value
plot(germs, lambdas, ylab='Population growth rate λ',
xlab='Germination rate; spring', col='blue')
abline(a=1, b=0)
From analytic solution:
# λ = 0.6561 + 3.8439 * germ
curve(0.6561 + 3.8439 * x, add=T,
from=min(germs), to=max(germs), col='red')
Both results met very well.
The plant:
plants <- seq(from=0, to=0.1, length.out=n)
for(i in 1:n) {
plant <- plants[i]
lambdas[i] <- lambda(A.spring())
}
plant <- 0.05 # restore the initial value
plot(plants, lambdas, ylab='Population growth rate λ',
xlab='Plant surviving rate; from germination to mature',
col='blue')
abline(a=1, b=0)
From analytic solution:
# λ = 0.45927 + 27 * plant
curve(0.45927 + 27 * x, add=T,
from=min(plants), to=max(plants), col='red')
Both results met very well.
The yield:
yields <- seq(from=0, to=200, length.out=n)
for(i in 1:n) {
yield <- yields[i]
lambdas[i] <- lambda(A.spring())
}
yield <- 100 # restore the initial value
plot(yields, lambdas, ylab='Population growth rate λ',
xlab='Seed production number; per matured plant',
col='blue')
abline(a=1, b=0)
From analytic solution:
# λ = 0.45927 + 0.0135 * yield
curve(0.45927 + 0.0135 * x, add=T,
from=min(yields), to=max(yields), col='red')
Both results met very well.
The seed and the germ:
n <- 64
lambdas <- matrix(nrow=n, ncol=n)
plant <- 0.05 # Plant surviving rate; from germination to mature
yield <- 100 # Seed production number; per matured plant
seeds <- seq(from=0, to=1, length.out=n)^2
germs <- seq(from=0, to=1, length.out=n)^2
for(ro in 1:n) for(co in 1:n) {
seed <- seeds[ro]
germ <- germs[co]
lambdas[ro, co] <- lambda(A.spring())
}
contour(x=seeds, y=germs, z=lambdas, main='λ',
xlab='Seed surviving rate; annual',
ylab='Germination rate; spring')
The seed and the plant:
n <- 64
lambdas <- matrix(nrow=n, ncol=n)
germ <- 0.3 # Germination rate; spring
yield <- 100 # Seed production number; per matured plant
seeds <- seq(from=0, to=1, length.out=n)^2
plants <- seq(from=0, to=0.3, length.out=n)^2
for(ro in 1:n) for(co in 1:n) {
seed <- seeds[ro]
plant <- plants[co]
lambdas[ro, co] <- lambda(A.spring())
}
contour(x=seeds, y=plants, z=lambdas,
xlab='Seed surviving rate; annual', main='λ',
ylab='Plant surviving rate; from germination to mature')
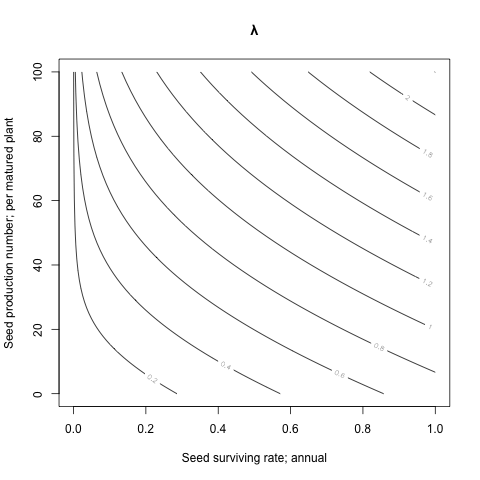
The seed and the yield:
n <- 64
lambdas <- matrix(nrow=n, ncol=n)
germ <- 0.3 # Germination rate; spring
plant <- 0.05 # Plant surviving rate; from germination to mature
seeds <- seq(from=0, to=1, length.out=n)^2
yields <- seq(from=0, to=sqrt(100), length.out=n)^2
for(ro in 1:n) for(co in 1:n) {
seed <- seeds[ro]
yield <- yields[co]
lambdas[ro, co] <- lambda(A.spring())
}
contour(x=seeds, y=yields, z=lambdas, main='λ',
xlab='Seed surviving rate; annual',
ylab='Seed production number; per matured plant')
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.