Sensitivity analysis for neural networks
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
I’ve made quite a few blog posts about neural networks and some of the diagnostic tools that can be used to ‘demystify’ the information contained in these models. Frankly, I’m kind of sick of writing about neural networks but I wanted to share one last tool I’ve implemented in R. I’m a strong believer that supervised neural networks can be used for much more than prediction, as is the common assumption by most researchers. I hope that my collection of posts, including this one, has shown the versatility of these models to develop inference into causation. To date, I’ve authored posts on visualizing neural networks, animating neural networks, and determining importance of model inputs. This post will describe a function for a sensitivity analysis of a neural network. Specifically, I will describe an approach to evaluate the form of the relationship of a response variable with the explanatory variables used in the model.
The general goal of a sensitivity analysis is similar to evaluating relative importance of explanatory variables, with a few important distinctions. For both analyses, we are interested in the relationships between explanatory and response variables as described by the model in the hope that the neural network has explained some real-world phenomenon. Using Garson’s algorithm,1 we can get an idea of the magnitude and sign of the relationship between variables relative to each other. Conversely, the sensitivity analysis allows us to obtain information about the form of the relationship between variables rather than a categorical description, such as variable x is positively and strongly related to y. For example, how does a response variable change in relation to increasing or decreasing values of a given explanatory variable? Is it a linear response, non-linear, uni-modal, no response, etc.? Furthermore, how does the form of the response change given values of the other explanatory variables in the model? We might expect that the relationship between a response and explanatory variable might differ given the context of the other explanatory variables (i.e., an interaction may be present). The sensitivity analysis can provide this information.
As with most of my posts, I’ve created the sensitivity analysis function using ideas from other people that are much more clever than me. I’ve simply converted these ideas into a useful form in R. Ultimate credit for the sensitivity analysis goes to Sovan Lek (and colleagues), who developed the approach in the mid-1990s. The ‘Lek-profile method’ is described briefly in Lek et al. 19962 and in more detail in Gevrey et al. 2003.3 I’ll provide a brief summary here since the method is pretty simple. In fact, the profile method can be extended to any statistical model and is not specific to neural networks, although it is one of few methods used to evaluate the latter. For any statistical model where multiple response variables are related to multiple explanatory variables, we choose one response and one explanatory variable. We obtain predictions of the response variable across the range of values for the given explanatory variable. All other explanatory variables are held constant at a given set of respective values (e.g., minimum, 20th percentile, maximum). The final product is a set of response curves for one response variable across the range of values for one explanatory variable, while holding all other explanatory variables constant. This is implemented in R by creating a matrix of values for explanatory variables where the number of rows is the number of observations and the number of columns is the number of explanatory variables. All explanatory variables are held at their mean (or other constant value) while the variable of interest is sequenced from its minimum to maximum value across the range of observations. This matrix (actually a data frame) is then used to predict values of the response variable from a fitted model object. This is repeated for different variables.
I’ll illustrate the function using simulated data, as I’ve done in previous posts. The exception here is that I’ll be using two response variables instead of one. The two response variables are linear combinations of eight explanatory variables, with random error components taken from a normal distribution. The relationships between the variables are determined by the arbitrary set of parameters (parms1 and parms2). The explanatory variables are partially correlated and taken from a multivariate normal distribution.
require(clusterGeneration)
require(nnet)
#define number of variables and observations
set.seed(2)
num.vars<-8
num.obs<-10000
#define correlation matrix for explanatory variables
#define actual parameter values
cov.mat<-genPositiveDefMat(num.vars,covMethod=c("unifcorrmat"))$Sigma
rand.vars<-mvrnorm(num.obs,rep(0,num.vars),Sigma=cov.mat)
parms1<-runif(num.vars,-10,10)
y1<-rand.vars %*% matrix(parms1) + rnorm(num.obs,sd=20)
parms2<-runif(num.vars,-10,10)
y2<-rand.vars %*% matrix(parms2) + rnorm(num.obs,sd=20)
#prep data and create neural network
rand.vars<-data.frame(rand.vars)
resp<-apply(cbind(y1,y2),2, function(y) (y-min(y))/(max(y)-min(y)))
resp<-data.frame(resp)
names(resp)<-c('Y1','Y2')
mod1<-nnet(rand.vars,resp,size=8,linout=T)
Here’s what the model looks like:
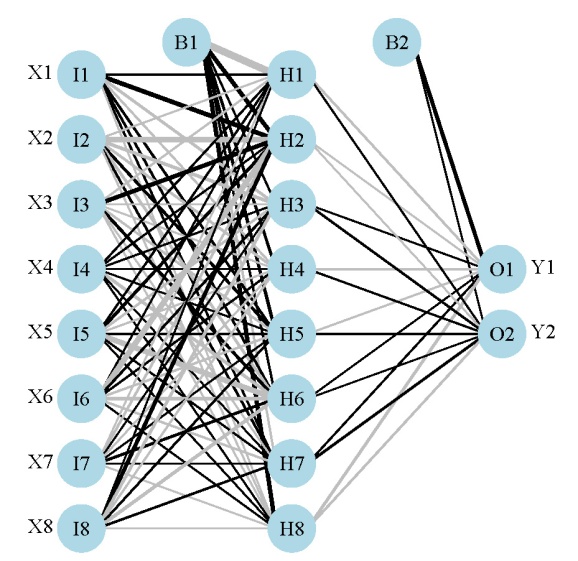
We’ve created a neural network that hopefully describes the relationship of two response variables with eight explanatory variables. The sensitivity analysis lets us visualize these relationships. The Lek profile function can be used once we have a neural network model in our workspace. The function is imported and used as follows:
source('https://gist.github.com/fawda123/6860630/raw/e50fc6ef30b8269660b4e65aeec7ce02beb9b551/lek_fun.r')
lek.fun(mod1)

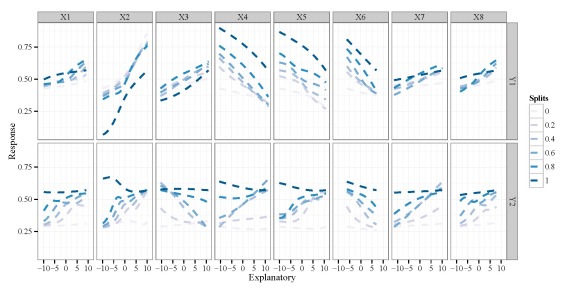
Each facet of the plot shows the bivariate relationship between one response variable and one explanatory variable. The multiple lines per plot indicate the change in the relationship when the other explanatory variables are held constant, in this case at their minimum, 20th, 40th, 60th, 80th, and maximum quantile values (the splits variable in the legend). Since our data were random we don’t necessarily care about the relationships, but you can see the wealth of information that could be provided by this plot if we don’t know the actual relationships between the variables.
The function takes the following arguments:
mod.in |
model object for input created from nnet function (nnet package) |
var.sens |
vector of character strings indicating the explanatory variables to evaluate, default NULL will evaluate all |
resp.name |
vector of character strings indicating the reponse variables to evaluate, default NULL will evaluate all |
steps |
numeric value indicating number of observations to evaluate for each explanatory variable from minimum to maximum value, default 100 |
split.vals |
numeric vector indicating quantile values at which to hold other explanatory variables constant |
val.out |
logical value indicating if actual sensitivity values are returned rather than a plot, default F |
By default, the function runs a sensitivity analysis for all variables. This creates a busy plot so we may want to look at specific variables of interest. Maybe we want to evaluate different quantile values as well. These options can be changed using the arguments.
lek.fun(mod1,var.sens=c('X2','X5'),split.vals=seq(0,1,by=0.05))

X2 and X5 and different quantile values for the remaining variables.
The function also returns a ggplot2 object that can be further modified. You may prefer a different theme, color, or line type, for example.
p1<-lek.fun(mod1)
class(p1)
# [1] "gg" "ggplot"
p1 +
theme_bw() +
scale_colour_brewer(palette="PuBu") +
scale_linetype_manual(values=rep('dashed',6)) +
scale_size_manual(values=rep(1,6))
ggplot options for the sensitivity analysis.
Finally, the actual values from the sensitivity analysis can be returned if you’d prefer that instead. The output is a data frame in long form that was created using melt.list from the reshape package for compatibility with ggplot2. The six columns indicate values for explanatory variables on the x-axes, names of the response variables, predicted values of the response variables, quantiles at which other explanatory variables were held constant, and names of the explanatory variables on the x-axes.
head(lek.fun(mod1,val.out=T)) # Explanatory resp.name Response Splits exp.name # 1 -9.825388 Y1 0.4285857 0 X1 # 2 -9.634531 Y1 0.4289905 0 X1 # 3 -9.443674 Y1 0.4293973 0 X1 # 4 -9.252816 Y1 0.4298058 0 X1 # 5 -9.061959 Y1 0.4302162 0 X1 # 6 -8.871102 Y1 0.4306284 0 X1
I mentioned earlier that the function is not unique to neural networks and can work with other models created in R. I haven’t done an extensive test of the function, but I’m fairly certain that it will work if the model object has a predict method (e.g., predict.lm). Here’s an example using the function to evaluate a multiple linear regression for one of the response variables.
mod2<-lm(Y1~.,data=cbind(resp[,'Y1',drop=F],rand.vars)) lek.fun(mod2)
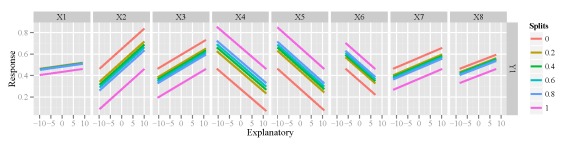
Y1 response variable.
This function has little relevance for conventional models like linear regression since a wealth of diagnostic tools are already available (e.g., effects plots, add/drop procedures, outlier tests, etc.). The application of the function to neural networks provides insight into the relationships described by the models, insights that to my knowledge, cannot be obtained using current tools in R. This post concludes my contribution of diagnostic tools for neural networks in R and I hope that they have been useful to some of you. I have spent the last year or so working with neural networks and my opinion of their utility is mixed. I see advantages in the use of highly flexible computer-based algorithms, although in most cases similar conclusions can be made using more conventional analyses. I suggest that neural networks only be used if there is an extremely high sample size and other methods have proven inconclusive. Feel free to voice your opinions or suggestions in the comments.
Cheers,
Marcus
1Garson GD. 1991. Interpreting neural network connection weights. Artificial Intelligence Expert. 6:46–51.
2Lek S, Delacoste M, Baran P, Dimopoulos I, Lauga J, Aulagnier S. 1996. Application of neural networks to modelling nonlinear relationships in Ecology. Ecological Modelling. 90:39-52.
3Gevrey M, Dimopoulos I, Lek S. 2003. Review and comparison of methods to study the contribution of variables in artificial neural network models. Ecological Modelling. 160:249-264.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
