How to make a scientific result disappear
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Nathan Danneman (a co-author and one of my graduate students from Emory) recently sent me a New Yorker article from 2010 about the “decline effect,” the tendency for initially promising scientific results to get smaller upon replication. Wikipedia can summarize the phenomenon as well as I can:
In his article, Lehrer gives several examples where the decline effect is allegedly showing. In the first example, the development of second generation anti-psychotic drugs, reveals that the first tests had demonstrated a dramatic decrease in the subjects’ psychiatric symptoms. However, after repeating tests this effect declined and in the end it was not possible to document that these drugs had any better effect than the first generation anti-psychotics.
Experiments done by Jonathan Schooler were trying to prove that people describing their memories were less able to remember them than people not describing their memories. His first experiments were positive, proving his theory about verbal overshadowing but repeated studies showed a significant declining effect.
In 1991, Danish zoologist Anders Møller discovered a connection between symmetry and sexual preference of females in nature. This sparked a huge interest in the topic and a lot of follow-up research was published. In three years following the original discovery, 90% of studies confirmed Møller’s hypothesis. However, the same outcome was published in just four out of eight research papers in 1995, and only a third in next three years.
Why would a treatment that shows a huge causal effect in an experiment seem to get weaker when that experiment is repeated later on? “‘This was profoundly frustrating,’ he [Schooler] says. ‘It was as if nature gave me this great result and then tried to take it back.’”
The cosmos may be indifferent to our plight, but I don’t think it’s actually vindictive (or at least does not express its malice through toying with our experimental results). PZ Myers proposes multiple, less vindictive explanations; two of them make a great deal of sense to me.
Regression to the mean: As the number of data points increases, we expect the average values to regress to the true mean…and since often the initial work is done on the basis of promising early results, we expect more data to even out a fortuitously significant early outcome.
The file drawer effect: Results that are not significant are hard to publish, and end up stashed away in a cabinet. However, as a result becomes established, contrary results become more interesting and publishable.
These are common, well-known and well-understood phenomena. But as far as I know, no one’s really tried to formally assess the impact of these phenomena or to propose any kind of diagnostic of how susceptible any particular result is to these threats to inference.
Let’s start with a simple example. Suppose that the data generating process is , where
. If we repeatedly generate data sets of size 1000 out of this DGP, run an appropriate linear model
, and save only those estimated coefficients that are statistically significant in a one-tailed test,
.
# what the distribution of statistically significant results looks like
set.seed(23409)
all.beta<-c()
sig.beta<-c()
for(i in 1:5000){
x<-runif(1000)
y<-0.5*x+rnorm(1000, mean=0, sd=4)
sig.beta[i]<-ifelse(summary(lm(y~x))$coefficients[2,3]>qnorm(0.95), summary(lm(y~x))$coefficients[2,1], NA)
all.beta[i]<-summary(lm(y~x))$coefficients[2,1]
}
hist(sig.beta, xlim=c(0,2.5), ylim=c(0, 400), xlab=expression(paste("Estimated Coefficient ", hat(beta))), main=c("Distribution of Statistically", "Significant Coefficients, beta = 0.5"))
abline(v=0.5, lty=2)
mean(sig.beta, na.rm=T)
And what do we get?
In short, we find that none of the statistically significant results are near the actual coefficient of 0.5. In fact, the statistically significant coefficients are biased upward (the mean coefficient is 1.20 1.008 in this simulation). This makes sense: only the largest slopes are capable of overcoming the intrinsic noise in the DGP and being detected at this sample size (1000).
What does this mean? Well… the estimator is not itself intrinsically biased: if you plotted all the coefficients from our 1000 simulated samples, they would be normally distributed around the true mean of 0.5 with appropriate variance. But we’re not talking about the distribution of an estimator given a true value, ; we’re talking about the distribution of scientifically notable, publishable results
. This is the distribution of results we expect to see in journal articles and in the media. And that distribution is biased because the scientific review process requires that results reach a certain signal-to-noise ratio (viz., a p-value smaller than 0.05) before they deserve scientific attention:
In short: when you look at what’s in journal articles or scientific magazines, you’re looking at results that are biased. Not that this is a terrible thing: we are imposing this bias so as to avoid printing a stream of chance results or reams of uninformative non-effects (do we need to know that the price of tea in China is not related to rainfall in London?).
In fact, given the size of the underlying effect, I can tell you precisely how large we should expect that bias to be. The analysis below is for a normally distributed with a standard error of 0.5.
# compute the mean estimate of beta hat given a true beta
# conditional on one-tailed 5% statistical significance
# assume a standard error of 0.5
set.seed(7712389)
samples<-rnorm(200000, mean=0, sd=0.5)
b<-seq(from=-1, to=3, by=0.05)
bias<-c()
mean.est<-c()
for(i in 1:length(b)){
mean.est[i]<-mean(ifelse(samples+b[i]>qnorm(0.95, sd=0.5), samples+b[i], NA), na.rm=T)
bias[i]<-mean.est[i]-b[i]
}
par(mar=c(5,5,4,2))
plot(bias~b, type="l", ylab=expression(paste("average bias (E[", hat(beta)-beta,"])")), xlab=expression(paste("true coefficient ", beta)), main=c("Bias in Expected Statisically", "Significant Regression Coefficients"))
So: the smaller the true coefficient, the larger we expect a statistically significant (and positive) estimate of that coefficient to be. The lesson is relatively straightforward: comparatively small relationships are very likely to be overestimated in the published literature, but larger relationships are more likely to be accurately estimated.
The bias plot I just produced is neat, but the x-axis shows the true beta value—which, of course, we cannot know in advance. It explains the overall pattern of overestimated effects in journal articles and medical studies, but doesn’t give us a way to assess any particular result. It would be better, I think, for us to have some guess about the probability that the coefficient we are seeing in a journal article is biased upward. I think we can get that, too.
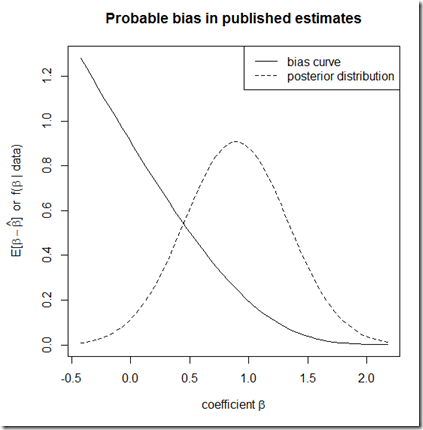
This block of code illustrates the gist of how I would proceed. I simulate a data set (of size 1000) out of the DGP ,
, and superimpose the posterior distribution of
(using the frequentist procedure, and therefore implying a flat prior) from that regression onto the bias curve computed for this application.
# now create an example of the averaged bias of
# an applied result
set.seed(389149)
x<-runif(1000)
y<-x+rnorm(1000, mean=0, sd=4)
summary(lm(y~x))
coefs<-summary(lm(y~x))$coefficients
samples<-rnorm(200000, mean=0, sd=coefs[2,2])
b<-seq(from=coefs[2,1]-3*coefs[2,2], to=coefs[2,1]+3*coefs[2,2], by=0.05)
bias<-c()
mean.est<-c()
for(i in 1:length(b)){
mean.est[i]<-mean(ifelse(samples+b[i]>qnorm(0.95, sd=coefs[2,2]), samples+b[i], NA), na.rm=T)
bias[i]<-mean.est[i]-b[i]
}
par(mar=c(5,5,4,2))
plot(bias~b, type="l", ylab=expression(paste("E[", beta-hat(beta), "] or f(", beta, " | data)")), xlab=expression(paste("coefficient ", beta)), main="Probable bias in published estimates")
lines(dnorm(b, mean=coefs[2,1], sd=coefs[2,2])~b, lty=2)
legend("topright", lty=c(1,2), legend=c("bias curve", "posterior distribution"))
The idea is to use the posterior distribution of as our best guess about the underlying state of the world, then to infer the expected bias in the published literature on the basis of this distribution. In principle, this can be done with any appropriate data set for the study of
; for example, I could imagine collecting data, running an analysis, finding a statistically insignificant
, and then constructing this plot to determine the distribution of this relationship that I will see published in extant or even future studies! But it’s probably more likely that someone will look at a published posterior, and then infer something about the likelihood that this published result overstates the actual effect.
That is, we would calculate
The inner integral is the expected bias of the estimated coefficient given its true value, and the outer integral calculates this expectation over the posterior.
I created some R code to calculate this expectation, and then applied it to the posterior distribution above.
# now compute the expected bias in the literature
# given a posterior
avg.bias<-function(b.est, se, seed=21381){
set.seed(seed)
samples<-rnorm(200000, mean=0, sd=se)
integrand<-function(x, b.est, se, samples){
out<-c()
for(i in 1:length(x)){
out[i]<-dnorm(x[i], mean=b.est, sd=se)*(mean(ifelse(samples+x[i]>qnorm(0.95, sd=se), samples+x[i], NA), na.rm=T)-x[i])
}
return(out)
}
return(integrate(f=integrand, lower=b.est-3*se, upper=b.est+3*se, b.est=b.est, se=se, samples=samples))
}
avg.bias(coefs[2,1], coefs[2,2])
avg.bias(2, coefs[2,2])
The result: an average bias of 0.352 0.299. What this tells us is that, given the range of true values of with which this result is consistent, we would on average expect it to overstate the true value by about
40% 33%.
Note again: the estimator itself is not biased, or is anything about the data set wrong. What this tells us is that, given the “gating” process in publishing that is imposed by statistical significance testing, this particular published result is likely to overstate the true value of . If all posteriors were equally likely to be published, the distribution of published estimates
would be symmetric about
and there would be no bias.
But not all results are equally susceptible: larger and more certain published results are more likely to accurately represent the actual value of because deviations of
that are smaller and larger than
are equally likely to be published, as we see by computing the expected bias for a coefficient of 2 instead of 0.895: the expected bias drops to
0.023 0.016.
So, why do effects weaken over time? As PZ Myers said, “However, as a result becomes established, contrary results become more interesting and publishable.” What that means is that the statistical significance threshold for publication disappears, and the publication distribution shifts from
What I hope that I’ve done here is to create a diagnostic tool to identify possibly problematic results before the replication happens.
Comments and criticisms welcome! I’m still figuring out whether any of this makes any sense.
[Update 2/27/2013, 10:27 AM CST: I made a couple of very small edits to the text, source code, and pictures. The text edits were minor LaTeX errors and clarifications. I also changed the code so as to reflect the standard error of the estimated result, rather than a close but not quite
[Update 2/27/2013, 5:10 PM CST: Minor bug in the avg.bias function fixed and results updated.]
[Update 2/27/2013, 11:08 PM CST: Minor bug in the initial example code; fixed and plot/results updated. See comments for bug report. Thanks, dogg13!]
[Update 2/28/2013, 9:45 AM CST: Changed plot 1 slightly for aesthetics.]
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.