[This article was first published on Freakonometrics - Tag - R-english, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here)
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
(bis repetita) Consider the following regression summary,Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
Call: lm(formula = y1 ~ x1) Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 3.0001 1.1247 2.667 0.02573 * x1 0.5001 0.1179 4.241 0.00217 ** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.237 on 9 degrees of freedom Multiple R-squared: 0.6665, Adjusted R-squared: 0.6295 F-statistic: 17.99 on 1 and 9 DF, p-value: 0.00217
obtained from
> data(anscombe) > reg1=lm(y1~x1,data=anscombe)
Can we say something if we look (only) at that output ? The intercept is significatively non-null, as well as the slope, the 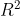

But it might be possible to have completely different kinds of patterns. Actually, four differents sets of data are coming from Anscombe (1973). And that all those datasets are somehow equivalent: the 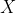
> apply(anscombe[,1:4],2,mean) x1 x2 x3 x4 9 9 9 9 > apply(anscombe[,1:4],2,var) x1 x2 x3 x4 11 11 11 11
and so are the 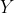
> apply(anscombe[,5:8],2,mean) y1 y2 y3 y4 7.500909 7.500909 7.500000 7.500909 > apply(anscombe[,5:8],2,var) y1 y2 y3 y4 4.127269 4.127629 4.122620 4.123249
Further, observe also that the correlation between the 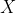
> cor(anscombe)[1:4,5:8] y1 y2 y3 y4 x1 0.8164205 0.8162365 0.8162867 -0.3140467 x2 0.8164205 0.8162365 0.8162867 -0.3140467 x3 0.8164205 0.8162365 0.8162867 -0.3140467 x4 -0.5290927 -0.7184365 -0.3446610 0.8165214 > diag(cor(anscombe)[1:4,5:8]) [1] 0.8164205 0.8162365 0.8162867 0.8165214
which yields the same regression line (intercept and slope)
> cbind(coef(reg1),coef(reg2),coef(reg3),coef(reg4)) [,1] [,2] [,3] [,4] (Intercept) 3.0000909 3.000909 3.0024545 3.0017273 x1 0.5000909 0.500000 0.4997273 0.4999091
But there is more. Much more. For instance, we always have the standard deviation for residuals
> c(summary(reg1)$sigma,summary(reg2)$sigma, + summary(reg3)$sigma,summary(reg4)$sigma) [1] 1.236603 1.237214 1.236311 1.235695
Thus, all regressions here have the same R2
> c(summary(reg1)$r.squared,summary(reg2)$r.squared, + summary(reg3)$r.squared,summary(reg4)$r.squared) [1] 0.6665425 0.6662420 0.6663240 0.6667073
Finally, Fisher’s F statistics is also (almost) the same.
+ c(summary(reg1)$fstatistic[1],summary(reg2)$fstatistic[1], + summary(reg3)$fstatistic[1],summary(reg4)$fstatistic[1]) value value value value 17.98994 17.96565 17.97228 18.00329
Thus, with the following datasets, we have the same prediction (and the same confidence intervals). Consider for instance the second dataset (the first one being mentioned above),
> reg2=lm(y2~x2,data=anscombe)
The output is here exactly the same as the one we had above
> summary(reg2) Call: lm(formula = y2 ~ x2, data = anscombe) Residuals: Min 1Q Median 3Q Max -1.9009 -0.7609 0.1291 0.9491 1.2691 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 3.001 1.125 2.667 0.02576 * x2 0.500 0.118 4.239 0.00218 ** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.237 on 9 degrees of freedom Multiple R-squared: 0.6662, Adjusted R-squared: 0.6292 F-statistic: 17.97 on 1 and 9 DF, p-value: 0.002179
Here, the perfect model is the one obtained with a quadratic regression.
> reg2b=lm(y2~x2+I(x2^2),data=anscombe) > summary(reg2b) Call: lm(formula = y2 ~ x2 + I(x2^2), data = anscombe) Residuals: Min 1Q Median 3Q Max -0.0013287 -0.0011888 -0.0006294 0.0008741 0.0023776 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -5.9957343 0.0043299 -1385 <2e-16 *** x2 2.7808392 0.0010401 2674 <2e-16 *** I(x2^2) -0.1267133 0.0000571 -2219 <2e-16 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 0.001672 on 8 degrees of freedom Multiple R-squared: 1, Adjusted R-squared: 1 F-statistic: 7.378e+06 on 2 and 8 DF, p-value: < 2.2e-16
Consider now the third one
> reg3=lm(y3~x3,data=anscombe)
i.e.
> summary(reg3) Call: lm(formula = y3 ~ x3, data = anscombe) Residuals: Min 1Q Median 3Q Max -1.1586 -0.6146 -0.2303 0.1540 3.2411 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 3.0025 1.1245 2.670 0.02562 * x3 0.4997 0.1179 4.239 0.00218 ** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.236 on 9 degrees of freedom Multiple R-squared: 0.6663, Adjusted R-squared: 0.6292 F-statistic: 17.97 on 1 and 9 DF, p-value: 0.002176
This time, the linear model could have been perfect. The problem is one outlier. If we remove it, we have
> reg3b=lm(y3~x3,data=anscombe[-3,]) > summary(reg3b) Call: lm(formula = y3 ~ x3, data = anscombe[-3, ]) Residuals: Min 1Q Median 3Q Max -0.0041558 -0.0022240 0.0000649 0.0018182 0.0050649 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.0056494 0.0029242 1370 <2e-16 *** x3 0.3453896 0.0003206 1077 <2e-16 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 0.003082 on 8 degrees of freedom Multiple R-squared: 1, Adjusted R-squared: 1 F-statistic: 1.161e+06 on 1 and 8 DF, p-value: < 2.2e-16
Finally consider
> reg4=lm(y4~x4,data=anscombe)
This time, there is an other kind of outlier, in
> summary(reg4) Call: lm(formula = y4 ~ x4, data = anscombe) Residuals: Min 1Q Median 3Q Max -1.751 -0.831 0.000 0.809 1.839 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 3.0017 1.1239 2.671 0.02559 * x4 0.4999 0.1178 4.243 0.00216 ** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.236 on 9 degrees of freedom Multiple R-squared: 0.6667, Adjusted R-squared: 0.6297 F-statistic: 18 on 1 and 9 DF, p-value: 0.002165
The graph is here
So clearly, looking at the summary of a regression does not tell us anything… This is why we do spend some time on diagnostic, looking at graphs with the errors (the graphs above could be obtained only with one explanatory variable, while errors can be studied in any dimension): everything can be seen on thise graphs. E.g. for the first dataset,
or the second one
the third one
or the fourth one,
To leave a comment for the author, please follow the link and comment on their blog: Freakonometrics - Tag - R-english.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
