Discriminating Between Iris Species
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
The Iris data set is a famous for its use to compare unsupervised classifiers.
The goal is to use information about flower characteristics to accurately classify the 3 species of Iris. We can look at scatter plots of the 4 variables in the data set and see that no single variable nor bivariate combination can achieve this.
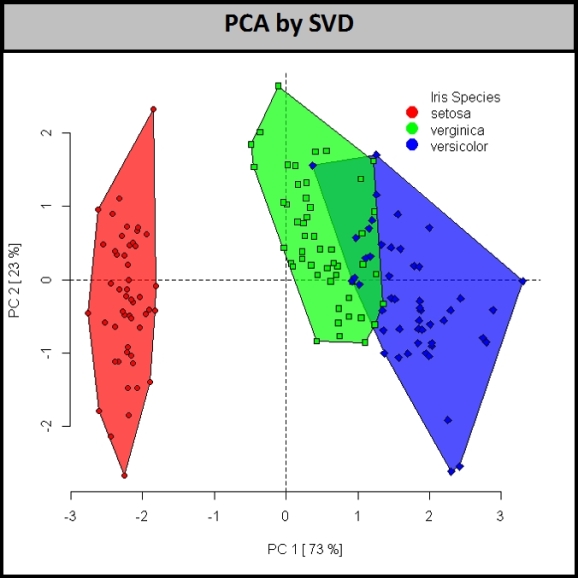
One approach to improve the separation between the two closely related Iris species, I.versicolor (blue) and I.virginica (green), is to use a combination of all 4 measurements, by constructing principal components (PCs).
Using the singular value decomposition to calculate PCs we see that the sample scores above are not resolved for the two species of interest.
Another approach is to use a supervised projection method like partial least squares (PLS), to identify Latent Variables (LVs) which are data projections similar to those of PCA, but which are also correlated with the species label. Interestingly this approach leads to a projection which changes the relative orientation of I. versicolor and I. verginica to I. setaosa. However, this supervised approach is not enough to identify a hyperplane of separation between all three species.
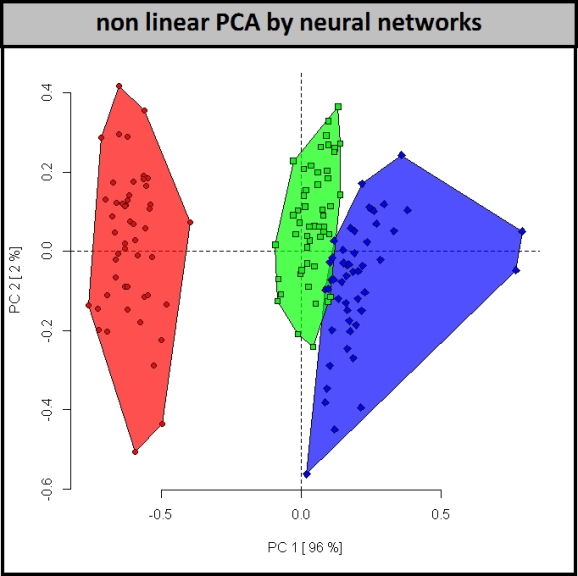
Non-linear PCA via neural networks can be used to identify the hypersurface of separation, shown above. Looking at the scores we can see that this approach is the most success for resolving the two closely related species. However, the loadings from this method, which help relate how the variables are combined achieve the classification, are impossible to interpret. In the case of the function used above(nlPca, pcaMethods R package) the loadings are literally NA.
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.