IPython Markdown Opportunities in IPython Notebooks and Rstudio
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.
One of the reasons I started working on the Wrangling F1 Data With R book was to see what the Rmd (RMarkdown) workflow was like. Rmd allows you to combine markdown and R code in the same document, as well as executing the code blocks and then displaying the results of that code execution inline in the output document.
As well as rendering to HTML, we can generate markdown (md is actually produced as the interim step to HTML creation), PDF output documents, etc etc.
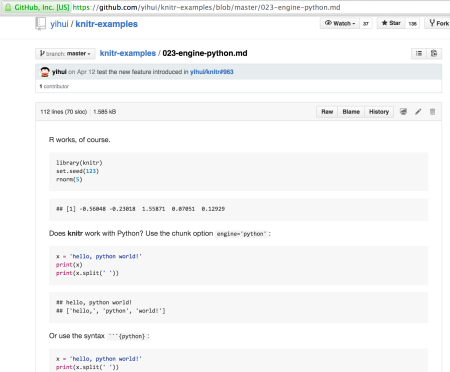
One thing I’d love to be able to do in the RStudio/RMarkdown environment is include – and execute – Python code. Does a web search to see what Python support there is in R… Ah, it seems it does it already… (how did I miss that?!)
ADDED: Unfortunately, it seems as if Python state is not persisted between separate python chunks – instead, each chunk is run as a one off python inline python command. However, it seems as if there could be a way round this, which is to use a persistent IPython session; and the knitron package looks like just the thing for supporting that.
So that means in RStudio, I could use knitr and Rmd to write a version of Wrangling F1 Data With RPython…
Of course, it would be nicer if I could write such a book in an everyday python environment – such as in an IPython notebook – that could also execute R code (just to be fair;-)
I know that we can already use cell magic to run R in a IPython notebook:
…so that’s that part of the equation.
And the notebooks do already allow us to mix markdown cells and code blocks/output. The default notebook presentation style is to show the code cells with the numbered In []: and Out []: block numbering, but it presumably only takes a small style extension or customisation to suppress that? And another small extension to add the ability to hide a code cell and just display the output?
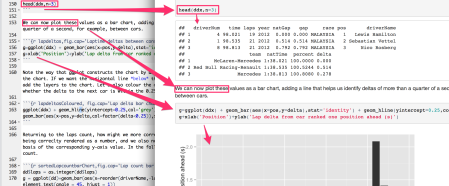
So what is it that (to my mind at least) makes RStudio a nicer writing environment? One reason is the ability to write the Rmarkdown simply as Rmarkdown in a simple text editor enviroment. Another is the ability to inline R code and display its output in-place.
Taking that second point first, the ability to do better inlining in IPython notebooks – it looks like this is just what the python-markdown extension seems to do:
But how about the ability to write some sort of pythonMarkdown and then open in a notebook? Something like ipymd, perhaps…?
What this seems to do is allow you to open an IPython-markdown document as an IPython notebook (in other words, it replaces the ipynb JSON document with an ipymd markdown document…). To support the document creation aspects better, we just need an exporter that removes the code block numbering and trivially allows code cells to be marked as hidden.
Now I wonder… what would it take to be able to open an Rmd document as an IPython notebook? Presumably just the ability to detect the code language, and then import the necessary magics to handle its execution? It’d be nice if it could cope with inline code, e.g. using the python-markdown magic too?
Exciting times could be ahead:-)
R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job.
Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.